Ron Daniel Jr
What's in a Measurement? Using GPT-3 on SemEval 2021 Task 8 -- MeasEval
Jun 28, 2021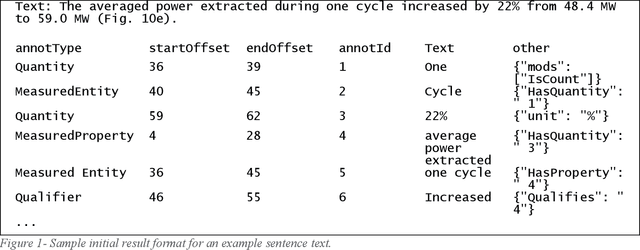
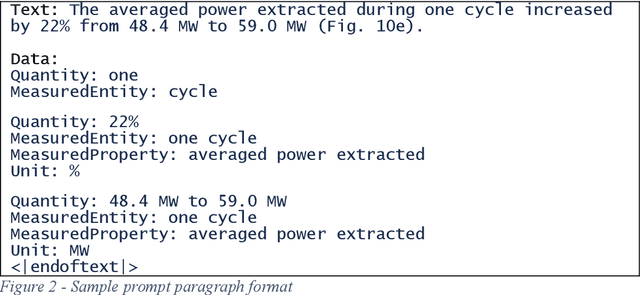


Abstract:In the summer of 2020 OpenAI released its GPT-3 autoregressive language model to much fanfare. While the model has shown promise on tasks in several areas, it has not always been clear when the results were cherry-picked or when they were the unvarnished output. We were particularly interested in what benefits GPT-3 could bring to the SemEval 2021 MeasEval task - identifying measurements and their associated attributes in scientific literature. We had already experimented with multi-turn questions answering as a solution to this task. We wanted to see if we could use GPT-3's few-shot learning capabilities to more easily develop a solution that would have better performance than our prior work. Unfortunately, we have not been successful in that effort. This paper discusses the approach we used, challenges we encountered, and results we observed. Some of the problems we encountered were simply due to the state of the art. For example, the limits on the size of the prompt and answer limited the amount of the training signal that could be offered. Others are more fundamental. We are unaware of generative models that excel in retaining factual information. Also, the impact of changes in the prompts is unpredictable, making it hard to reliably improve performance.
BioFLAIR: Pretrained Pooled Contextualized Embeddings for Biomedical Sequence Labeling Tasks
Aug 13, 2019
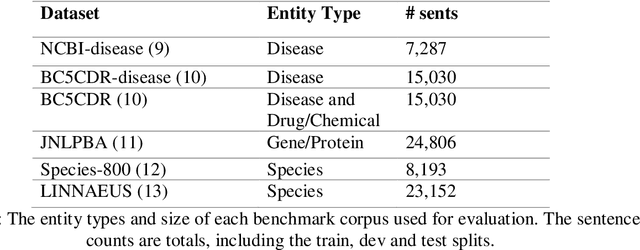
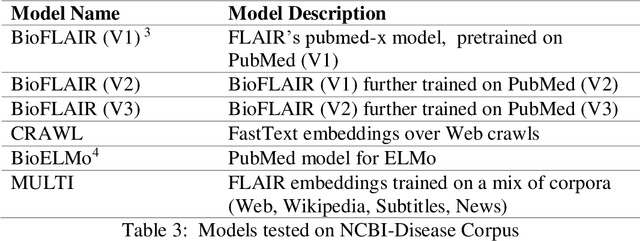
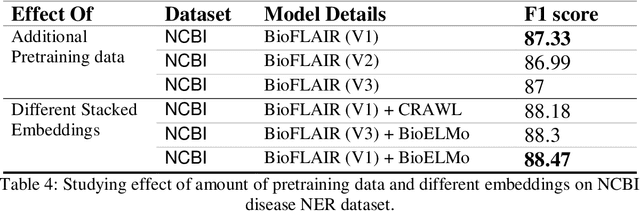
Abstract:Biomedical Named Entity Recognition (NER) is a challenging problem in biomedical information processing due to the widespread ambiguity of out of context terms and extensive lexical variations. Performance on bioNER benchmarks continues to improve due to advances like BERT, GPT, and XLNet. FLAIR (1) is an alternative embedding model which is less computationally intensive than the others mentioned. We test FLAIR and its pretrained PubMed embeddings (which we term BioFLAIR) on a variety of bio NER tasks and compare those with results from BERT-type networks. We also investigate the effects of a small amount of additional pretraining on PubMed content, and of combining FLAIR and ELMO models. We find that with the provided embeddings, FLAIR performs on-par with the BERT networks - even establishing a new state of the art on one benchmark. Additional pretraining did not provide a clear benefit, although this might change with even more pretraining being done. Stacking the FLAIR embeddings with others typically does provide a boost in the benchmark results.
Open Information Extraction on Scientific Text: An Evaluation
Jun 04, 2018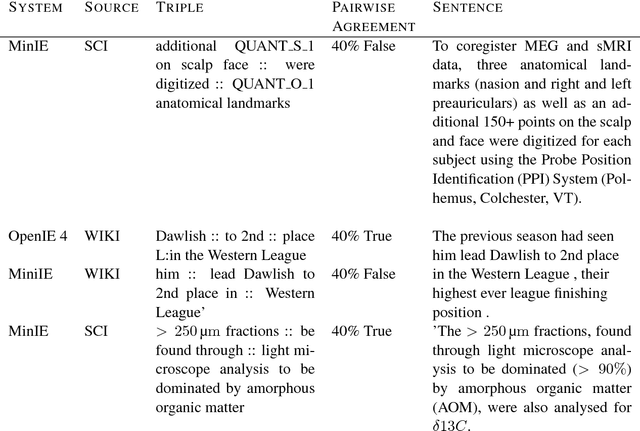
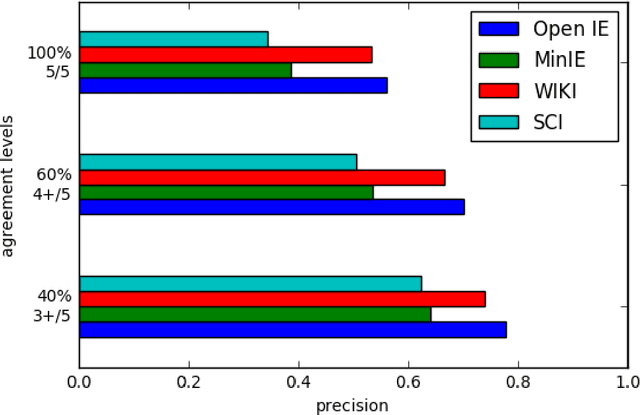
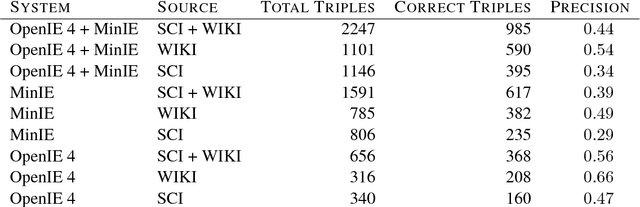
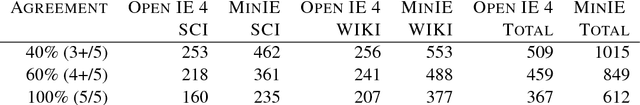
Abstract:Open Information Extraction (OIE) is the task of the unsupervised creation of structured information from text. OIE is often used as a starting point for a number of downstream tasks including knowledge base construction, relation extraction, and question answering. While OIE methods are targeted at being domain independent, they have been evaluated primarily on newspaper, encyclopedic or general web text. In this article, we evaluate the performance of OIE on scientific texts originating from 10 different disciplines. To do so, we use two state-of-the-art OIE systems applying a crowd-sourcing approach. We find that OIE systems perform significantly worse on scientific text than encyclopedic text. We also provide an error analysis and suggest areas of work to reduce errors. Our corpus of sentences and judgments are made available.
* 10 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge