Savitha Srinivasan
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024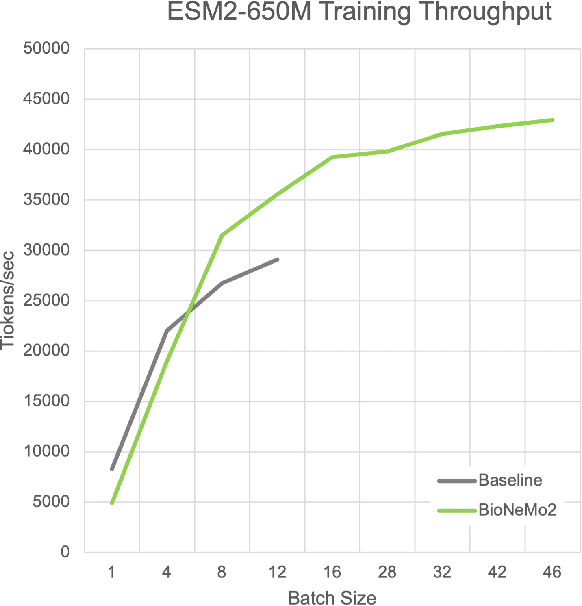
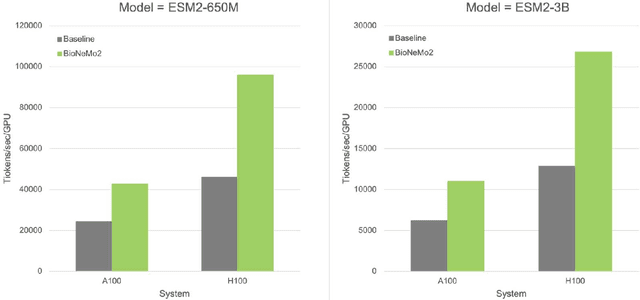
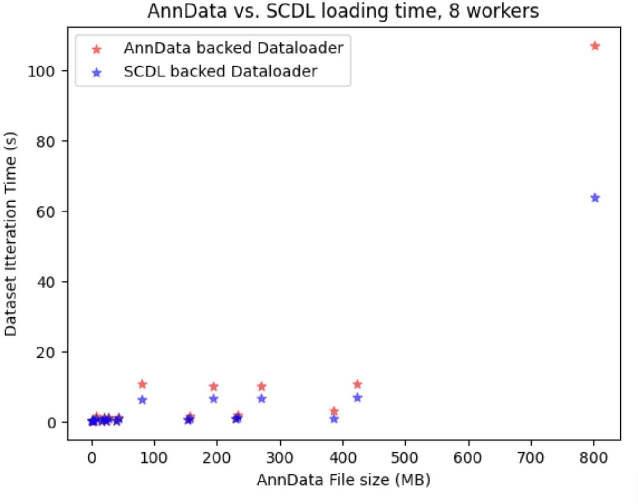
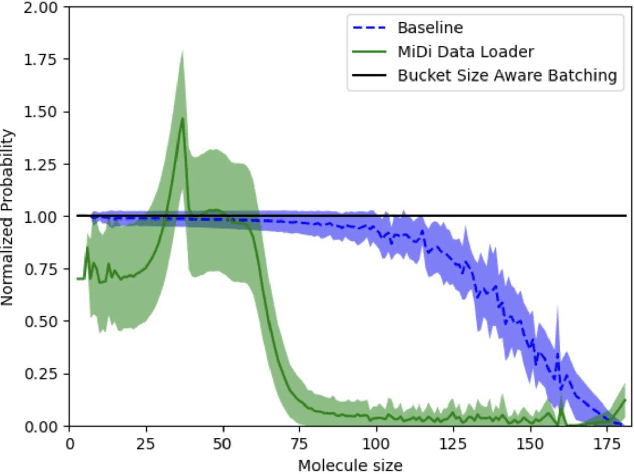
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
Domain Adaptation for Object Detection using SE Adaptors and Center Loss
May 25, 2022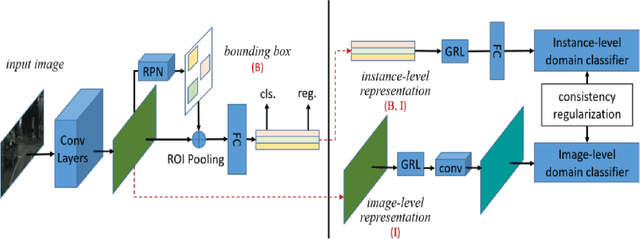
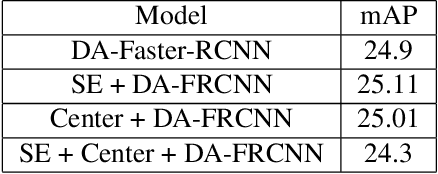
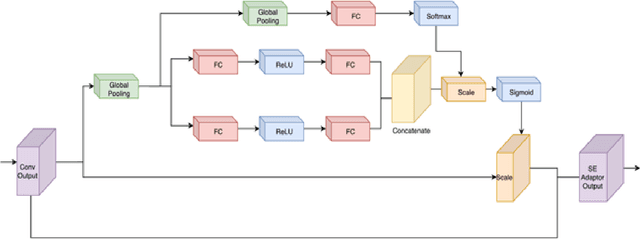
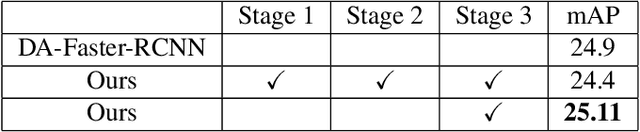
Abstract:Despite growing interest in object detection, very few works address the extremely practical problem of cross-domain robustness especially for automative applications. In order to prevent drops in performance due to domain shift, we introduce an unsupervised domain adaptation method built on the foundation of faster-RCNN with two domain adaptation components addressing the shift at the instance and image levels respectively and apply a consistency regularization between them. We also introduce a family of adaptation layers that leverage the squeeze excitation mechanism called SE Adaptors to improve domain attention and thus improves performance without any prior requirement of knowledge of the new target domain. Finally, we incorporate a center loss in the instance and image level representations to improve the intra-class variance. We report all results with Cityscapes as our source domain and Foggy Cityscapes as the target domain outperforming previous baselines.
Structure Aware and Class Balanced 3D Object Detection on nuScenes Dataset
May 25, 2022Abstract:3-D object detection is pivotal for autonomous driving. Point cloud based methods have become increasingly popular for 3-D object detection, owing to their accurate depth information. NuTonomy's nuScenes dataset greatly extends commonly used datasets such as KITTI in size, sensor modalities, categories, and annotation numbers. However, it suffers from severe class imbalance. The Class-balanced Grouping and Sampling paper addresses this issue and suggests augmentation and sampling strategy. However, the localization precision of this model is affected by the loss of spatial information in the downscaled feature maps. We propose to enhance the performance of the CBGS model by designing an auxiliary network, that makes full use of the structure information of the 3D point cloud, in order to improve the localization accuracy. The detachable auxiliary network is jointly optimized by two point-level supervisions, namely foreground segmentation and center estimation. The auxiliary network does not introduce any extra computation during inference, since it can be detached at test time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge