Sandeep Ayyar
Towards FAIR protocols and workflows: The OpenPREDICT case study
Nov 20, 2019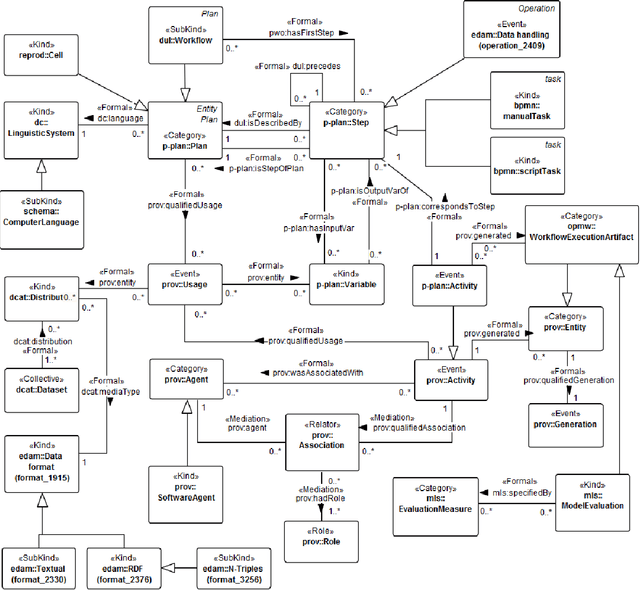
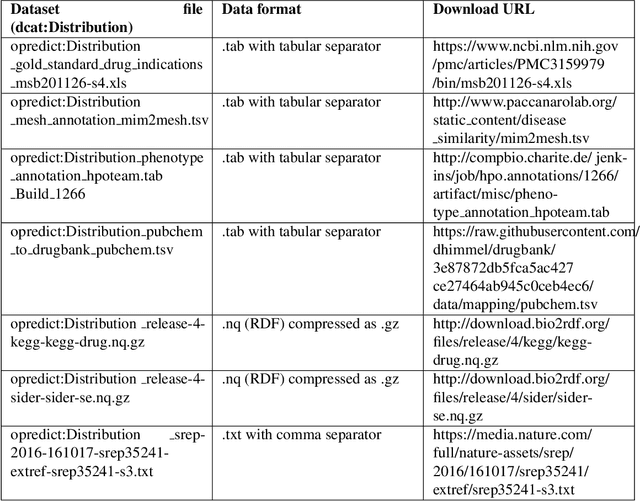
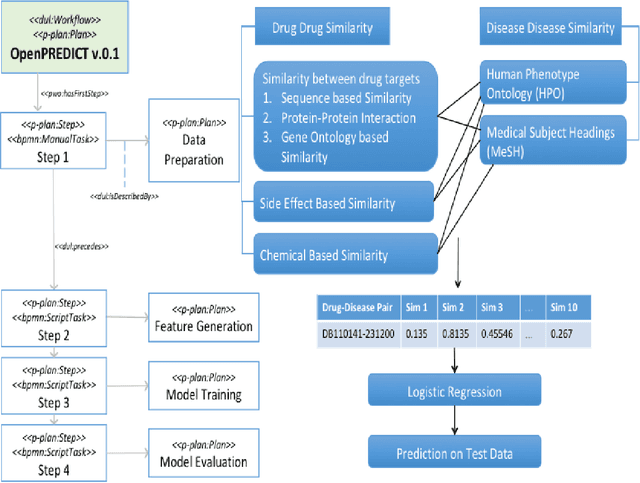
Abstract:It is essential for the advancement of science that scientists and researchers share, reuse and reproduce workflows and protocols used by others. The FAIR principles are a set of guidelines that aim to maximize the value and usefulness of research data, and emphasize a number of important points regarding the means by which digital objects are found and reused by others. The question of how to apply these principles not just to the static input and output data but also to the dynamic workflows and protocols that consume and produce them is still under debate and poses a number of challenges. In this paper we describe our inclusive and overarching approach to apply the FAIR principles to workflows and protocols and demonstrate its benefits. We apply and evaluate our approach on a case study that consists of making the PREDICT workflow, a highly cited drug repurposing workflow, open and FAIR. This includes FAIRification of the involved datasets, as well as applying semantic technologies to represent and store data about the detailed versions of the general protocol, of the concrete workflow instructions, and of their execution traces. A semantic model was proposed to better address these specific requirements and were evaluated by answering competency questions. This semantic model consists of classes and relations from a number of existing ontologies, including Workflow4ever, PROV, EDAM, and BPMN. This allowed us then to formulate and answer new kinds of competency questions. Our evaluation shows the high degree to which our FAIRified OpenPREDICT workflow now adheres to the FAIR principles and the practicality and usefulness of being able to answer our new competency questions.
Segmental Convolutional Neural Networks for Detection of Cardiac Abnormality With Noisy Heart Sound Recordings
Dec 06, 2016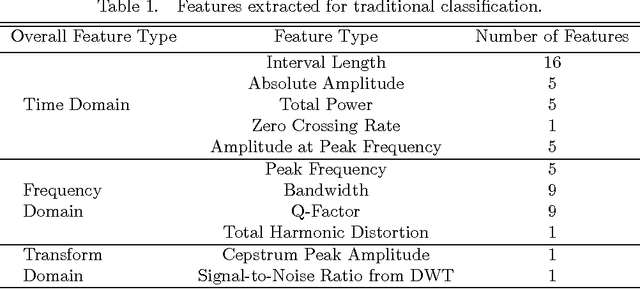
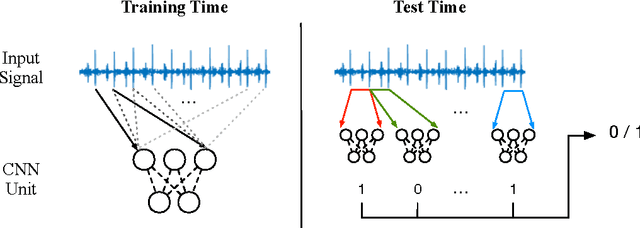
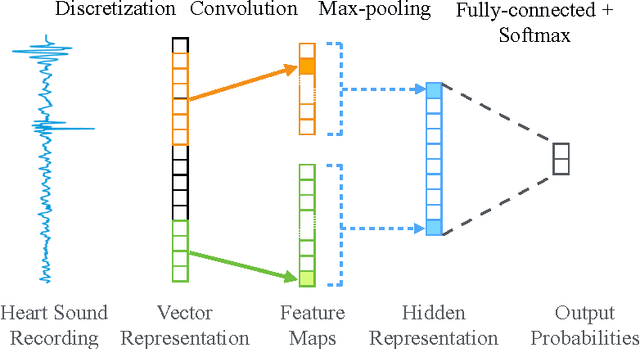
Abstract:Heart diseases constitute a global health burden, and the problem is exacerbated by the error-prone nature of listening to and interpreting heart sounds. This motivates the development of automated classification to screen for abnormal heart sounds. Existing machine learning-based systems achieve accurate classification of heart sound recordings but rely on expert features that have not been thoroughly evaluated on noisy recordings. Here we propose a segmental convolutional neural network architecture that achieves automatic feature learning from noisy heart sound recordings. Our experiments show that our best model, trained on noisy recording segments acquired with an existing hidden semi-markov model-based approach, attains a classification accuracy of 87.5% on the 2016 PhysioNet/CinC Challenge dataset, compared to the 84.6% accuracy of the state-of-the-art statistical classifier trained and evaluated on the same dataset. Our results indicate the potential of using neural network-based methods to increase the accuracy of automated classification of heart sound recordings for improved screening of heart diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge