Sabrina Guettes
A Comparison of Relaxations of Multiset Cannonical Correlation Analysis and Applications
Feb 05, 2013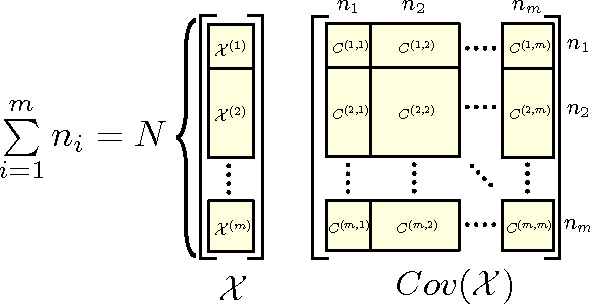
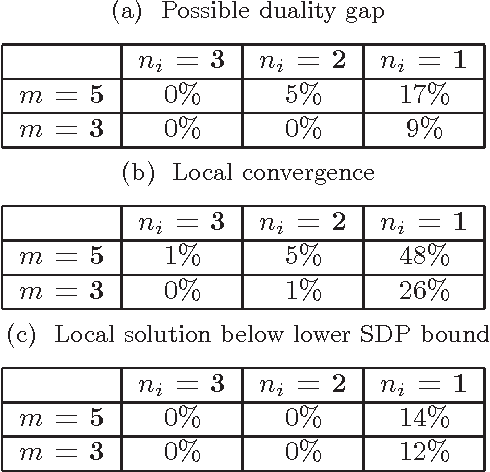
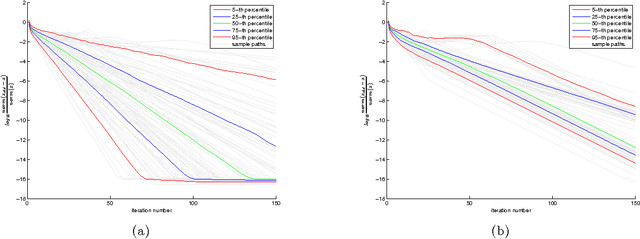
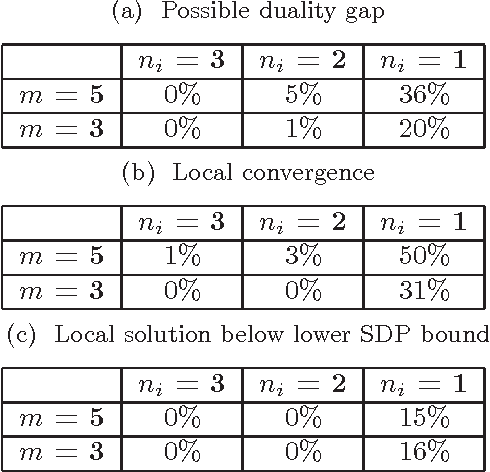
Abstract:Canonical correlation analysis is a statistical technique that is used to find relations between two sets of variables. An important extension in pattern analysis is to consider more than two sets of variables. This problem can be expressed as a quadratically constrained quadratic program (QCQP), commonly referred to Multi-set Canonical Correlation Analysis (MCCA). This is a non-convex problem and so greedy algorithms converge to local optima without any guarantees on global optimality. In this paper, we show that despite being highly structured, finding the optimal solution is NP-Hard. This motivates our relaxation of the QCQP to a semidefinite program (SDP). The SDP is convex, can be solved reasonably efficiently and comes with both absolute and output-sensitive approximation quality. In addition to theoretical guarantees, we do an extensive comparison of the QCQP method and the SDP relaxation on a variety of synthetic and real world data. Finally, we present two useful extensions: we incorporate kernel methods and computing multiple sets of canonical vectors.
Applications of Machine Learning Methods to Quantifying Phenotypic Traits that Distinguish the Wild Type from the Mutant Arabidopsis Thaliana Seedlings during Root Gravitropism
Aug 31, 2010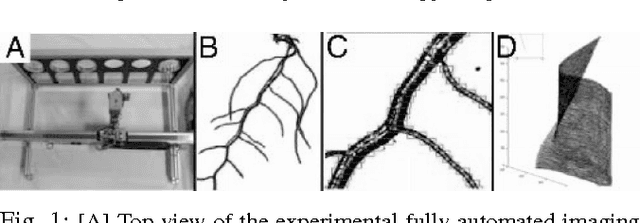

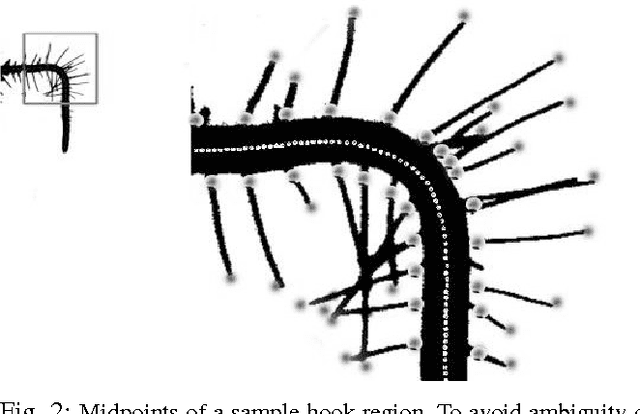
Abstract:Post-genomic research deals with challenging problems in screening genomes of organisms for particular functions or potential for being the targets of genetic engineering for desirable biological features. 'Phenotyping' of wild type and mutants is a time-consuming and costly effort by many individuals. This article is a preliminary progress report in research on large-scale automation of phenotyping steps (imaging, informatics and data analysis) needed to study plant gene-proteins networks that influence growth and development of plants. Our results undermine the significance of phenotypic traits that are implicit in patterns of dynamics in plant root response to sudden changes of its environmental conditions, such as sudden re-orientation of the root tip against the gravity vector. Including dynamic features besides the common morphological ones has paid off in design of robust and accurate machine learning methods to automate a typical phenotyping scenario, i.e. to distinguish the wild type from the mutants.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge