Ruiwei Feng
AGMI: Attention-Guided Multi-omics Integration for Drug Response Prediction with Graph Neural Networks
Jan 10, 2022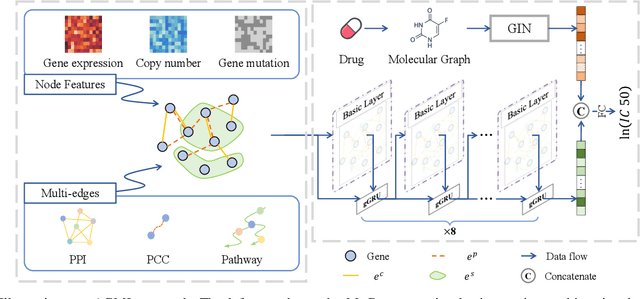
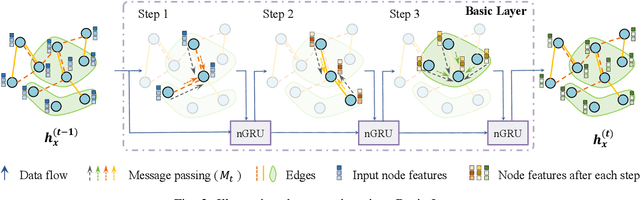
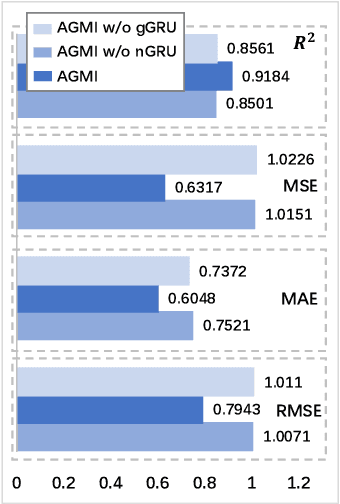
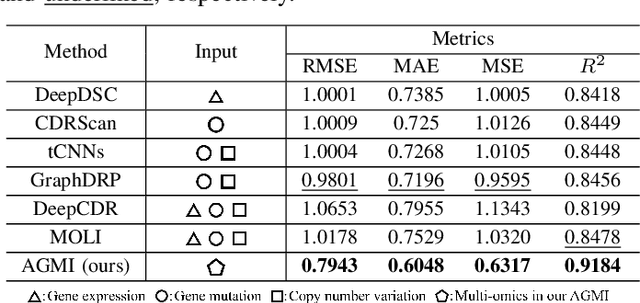
Abstract:Accurate drug response prediction (DRP) is a crucial yet challenging task in precision medicine. This paper presents a novel Attention-Guided Multi-omics Integration (AGMI) approach for DRP, which first constructs a Multi-edge Graph (MeG) for each cell line, and then aggregates multi-omics features to predict drug response using a novel structure, called Graph edge-aware Network (GeNet). For the first time, our AGMI approach explores gene constraint based multi-omics integration for DRP with the whole-genome using GNNs. Empirical experiments on the CCLE and GDSC datasets show that our AGMI largely outperforms state-of-the-art DRP methods by 8.3%--34.2% on four metrics. Our data and code are available at https://github.com/yivan-WYYGDSG/AGMI.
Doctor Imitator: A Graph-based Bone Age Assessment Framework Using Hand Radiographs
Feb 10, 2021



Abstract:Bone age assessment is challenging in clinical practice due to the complicated bone age assessment process. Current automatic bone age assessment methods were designed with rare consideration of the diagnostic logistics and thus may yield certain uninterpretable hidden states and outputs. Consequently, doctors can find it hard to cooperate with such models harmoniously because it is difficult to check the correctness of the model predictions. In this work, we propose a new graph-based deep learning framework for bone age assessment with hand radiographs, called Doctor Imitator (DI). The architecture of DI is designed to learn the diagnostic logistics of doctors using the scoring methods (e.g., the Tanner-Whitehouse method) for bone age assessment. Specifically, the convolutions of DI capture the local features of the anatomical regions of interest (ROIs) on hand radiographs and predict the ROI scores by our proposed Anatomy-based Group Convolution, summing up for bone age prediction. Besides, we develop a novel Dual Graph-based Attention module to compute patient-specific attention for ROI features and context attention for ROI scores. As far as we know, DI is the first automatic bone age assessment framework following the scoring methods without fully supervised hand radiographs. Experiments on hand radiographs with only bone age supervision verify that DI can achieve excellent performance with sparse parameters and provide more interpretability.
Flow-Mixup: Classifying Multi-labeled Medical Images with Corrupted Labels
Feb 09, 2021



Abstract:In clinical practice, medical image interpretation often involves multi-labeled classification, since the affected parts of a patient tend to present multiple symptoms or comorbidities. Recently, deep learning based frameworks have attained expert-level performance on medical image interpretation, which can be attributed partially to large amounts of accurate annotations. However, manually annotating massive amounts of medical images is impractical, while automatic annotation is fast but imprecise (possibly introducing corrupted labels). In this work, we propose a new regularization approach, called Flow-Mixup, for multi-labeled medical image classification with corrupted labels. Flow-Mixup guides the models to capture robust features for each abnormality, thus helping handle corrupted labels effectively and making it possible to apply automatic annotation. Specifically, Flow-Mixup decouples the extracted features by adding constraints to the hidden states of the models. Also, Flow-Mixup is more stable and effective comparing to other known regularization methods, as shown by theoretical and empirical analyses. Experiments on two electrocardiogram datasets and a chest X-ray dataset containing corrupted labels verify that Flow-Mixup is effective and insensitive to corrupted labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge