Roberto Ardon
Registration of Longitudinal Liver Examinations for Tumor Progress Assessment
Jan 24, 2025Abstract:Assessing cancer progression in liver CT scans is a clinical challenge, requiring a comparison of scans at different times for the same patient. Practitioners must identify existing tumors, compare them with prior exams, identify new tumors, and evaluate overall disease evolution. This process is particularly complex in liver examinations due to misalignment between exams caused by several factors. Indeed, longitudinal liver examinations can undergo different non-pathological and pathological changes due to non-rigid deformations, the appearance or disappearance of pathologies, and other variations. In such cases, existing registration approaches, mainly based on intrinsic features may distort tumor regions, biasing the tumor progress evaluation step and the corresponding diagnosis. This work proposes a registration method based only on geometrical and anatomical information from liver segmentation, aimed at aligning longitudinal liver images for aided diagnosis. The proposed method is trained and tested on longitudinal liver CT scans, with 317 patients for training and 53 for testing. Our experimental results support our claims by showing that our method is better than other registration techniques by providing a smoother deformation while preserving the tumor burden (total volume of tissues considered as tumor) within the volume. Qualitative results emphasize the importance of smooth deformations in preserving tumor appearance.
Decoupled conditional contrastive learning with variable metadata for prostate lesion detection
Aug 18, 2023



Abstract:Early diagnosis of prostate cancer is crucial for efficient treatment. Multi-parametric Magnetic Resonance Images (mp-MRI) are widely used for lesion detection. The Prostate Imaging Reporting and Data System (PI-RADS) has standardized interpretation of prostate MRI by defining a score for lesion malignancy. PI-RADS data is readily available from radiology reports but is subject to high inter-reports variability. We propose a new contrastive loss function that leverages weak metadata with multiple annotators per sample and takes advantage of inter-reports variability by defining metadata confidence. By combining metadata of varying confidence with unannotated data into a single conditional contrastive loss function, we report a 3% AUC increase on lesion detection on the public PI-CAI challenge dataset. Code is available at: https://github.com/camilleruppli/decoupled_ccl
Optimizing transformations for contrastive learning in a differentiable framework
Jul 27, 2022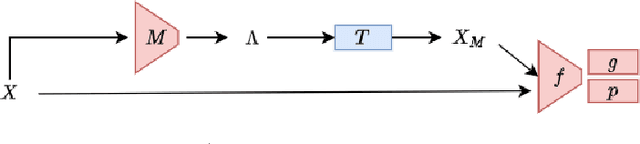
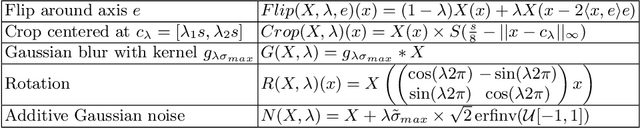
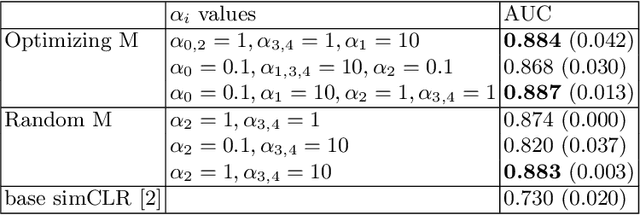
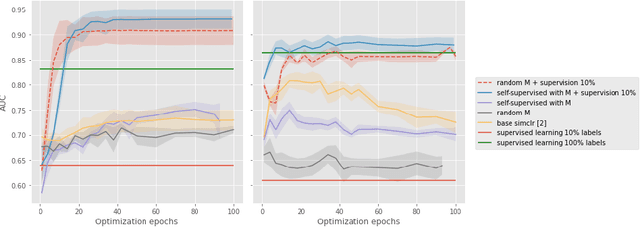
Abstract:Current contrastive learning methods use random transformations sampled from a large list of transformations, with fixed hyperparameters, to learn invariance from an unannotated database. Following previous works that introduce a small amount of supervision, we propose a framework to find optimal transformations for contrastive learning using a differentiable transformation network. Our method increases performances at low annotated data regime both in supervision accuracy and in convergence speed. In contrast to previous work, no generative model is needed for transformation optimization. Transformed images keep relevant information to solve the supervised task, here classification. Experiments were performed on 34000 2D slices of brain Magnetic Resonance Images and 11200 chest X-ray images. On both datasets, with 10% of labeled data, our model achieves better performances than a fully supervised model with 100% labels.
Leveraging Conditional Generative Models in a General Explanation Framework of Classifier Decisions
Jun 21, 2021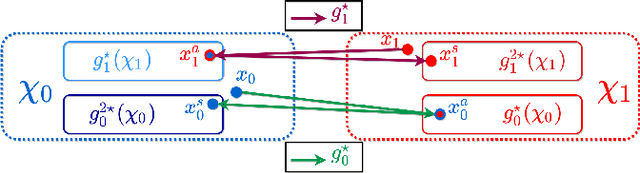
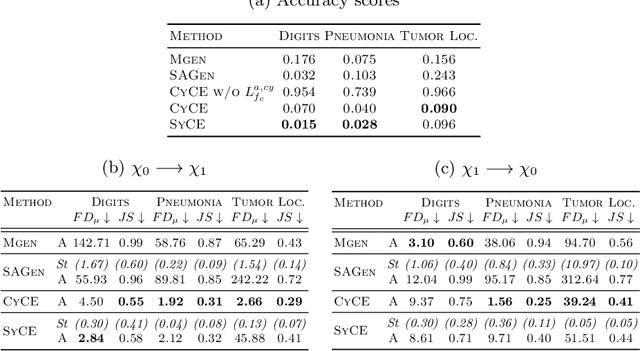
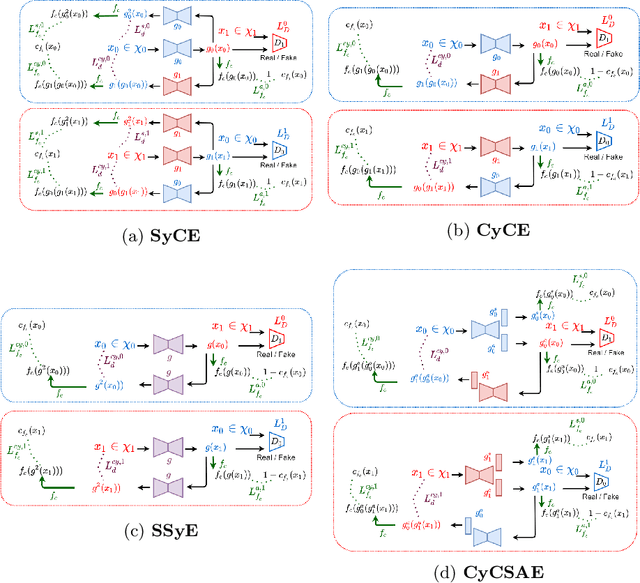
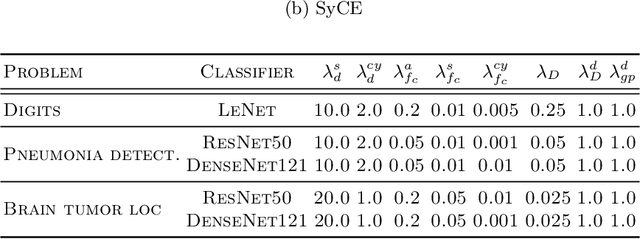
Abstract:Providing a human-understandable explanation of classifiers' decisions has become imperative to generate trust in their use for day-to-day tasks. Although many works have addressed this problem by generating visual explanation maps, they often provide noisy and inaccurate results forcing the use of heuristic regularization unrelated to the classifier in question. In this paper, we propose a new general perspective of the visual explanation problem overcoming these limitations. We show that visual explanation can be produced as the difference between two generated images obtained via two specific conditional generative models. Both generative models are trained using the classifier to explain and a database to enforce the following properties: (i) All images generated by the first generator are classified similarly to the input image, whereas the second generator's outputs are classified oppositely. (ii) Generated images belong to the distribution of real images. (iii) The distances between the input image and the corresponding generated images are minimal so that the difference between the generated elements only reveals relevant information for the studied classifier. Using symmetrical and cyclic constraints, we present two different approximations and implementations of the general formulation. Experimentally, we demonstrate significant improvements w.r.t the state-of-the-art on three different public data sets. In particular, the localization of regions influencing the classifier is consistent with human annotations.
Combining Similarity and Adversarial Learning to Generate Visual Explanation: Application to Medical Image Classification
Dec 14, 2020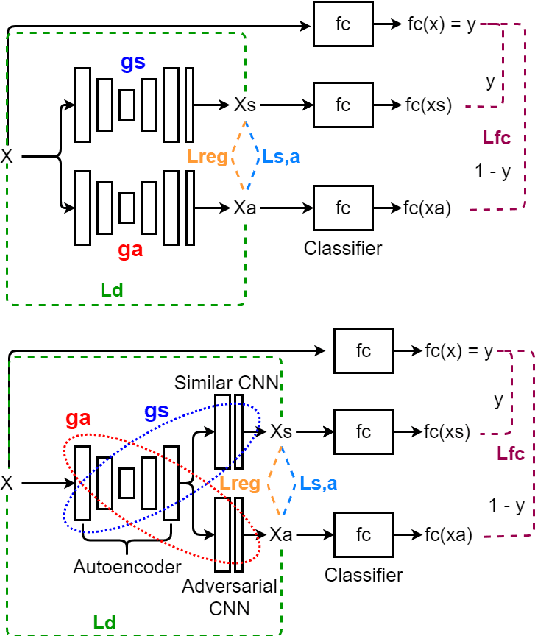
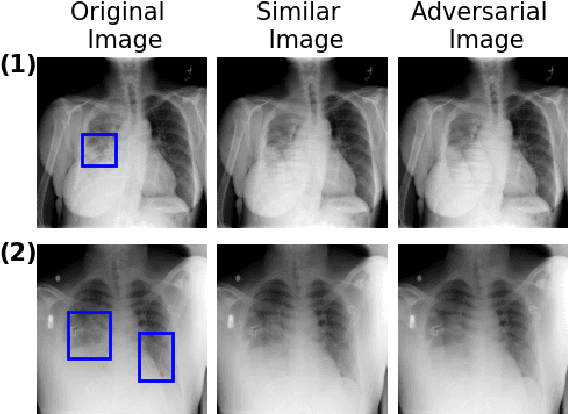
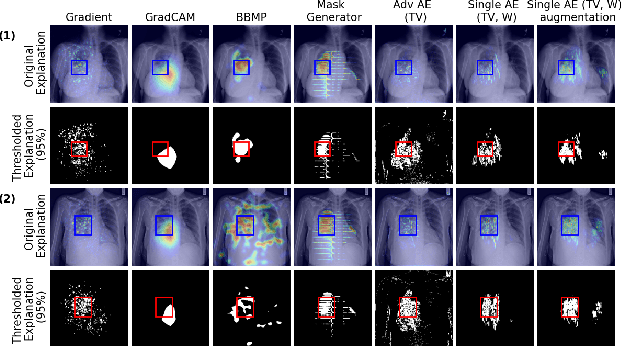
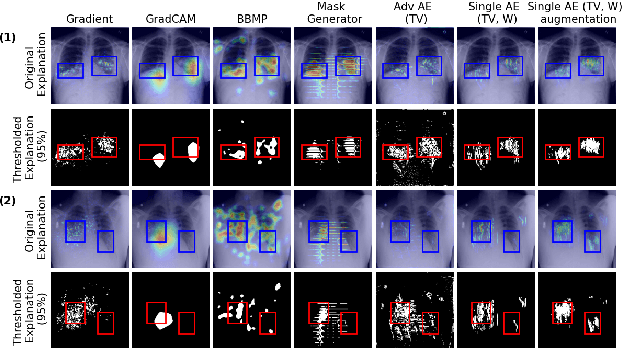
Abstract:Explaining decisions of black-box classifiers is paramount in sensitive domains such as medical imaging since clinicians confidence is necessary for adoption. Various explanation approaches have been proposed, among which perturbation based approaches are very promising. Within this class of methods, we leverage a learning framework to produce our visual explanations method. From a given classifier, we train two generators to produce from an input image the so called similar and adversarial images. The similar image shall be classified as the input image whereas the adversarial shall not. Visual explanation is built as the difference between these two generated images. Using metrics from the literature, our method outperforms state-of-the-art approaches. The proposed approach is model-agnostic and has a low computation burden at prediction time. Thus, it is adapted for real-time systems. Finally, we show that random geometric augmentations applied to the original image play a regularization role that improves several previously proposed explanation methods. We validate our approach on a large chest X-ray database.
Classification of MRI data using Deep Learning and Gaussian Process-based Model Selection
Jan 16, 2017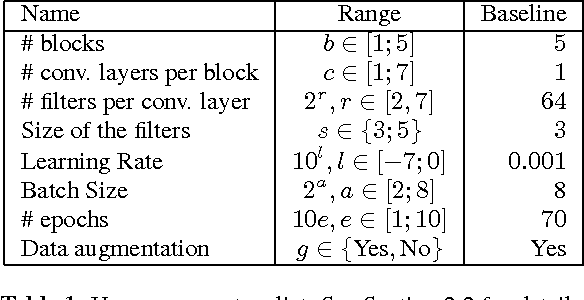
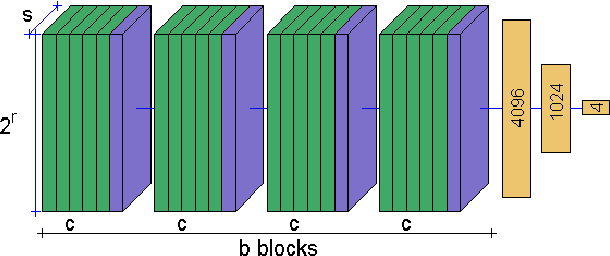
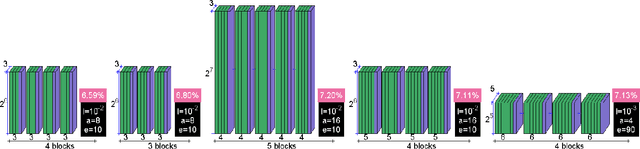
Abstract:The classification of MRI images according to the anatomical field of view is a necessary task to solve when faced with the increasing quantity of medical images. In parallel, advances in deep learning makes it a suitable tool for computer vision problems. Using a common architecture (such as AlexNet) provides quite good results, but not sufficient for clinical use. Improving the model is not an easy task, due to the large number of hyper-parameters governing both the architecture and the training of the network, and to the limited understanding of their relevance. Since an exhaustive search is not tractable, we propose to optimize the network first by random search, and then by an adaptive search based on Gaussian Processes and Probability of Improvement. Applying this method on a large and varied MRI dataset, we show a substantial improvement between the baseline network and the final one (up to 20\% for the most difficult classes).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge