Robert Clarke
Unsupervised deconvolution of dynamic imaging reveals intratumor vascular heterogeneity
Sep 04, 2014Abstract:Intratumor heterogeneity is often manifested by vascular compartments with distinct pharmacokinetics that cannot be resolved directly by in vivo dynamic imaging. We developed tissue-specific compartment modeling (TSCM), an unsupervised computational method of deconvolving dynamic imaging series from heterogeneous tumors that can improve vascular phenotyping in many biological contexts. Applying TSCM to dynamic contrast-enhanced MRI of breast cancers revealed characteristic intratumor vascular heterogeneity and therapeutic responses that were otherwise undetectable.
A feasible roadmap for unsupervised deconvolution of two-source mixed gene expressions
Oct 25, 2013
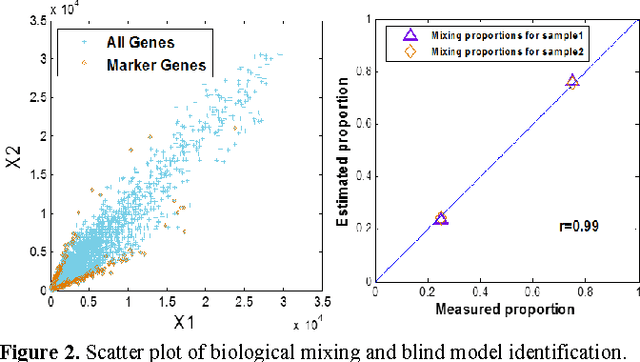


Abstract:Tissue heterogeneity is a major confounding factor in studying individual populations that cannot be resolved directly by global profiling. Experimental solutions to mitigate tissue heterogeneity are expensive, time consuming, inapplicable to existing data, and may alter the original gene expression patterns. Here we ask whether it is possible to deconvolute two-source mixed expressions (estimating both proportions and cell-specific profiles) from two or more heterogeneous samples without requiring any prior knowledge. Supported by a well-grounded mathematical framework, we argue that both constituent proportions and cell-specific expressions can be estimated in a completely unsupervised mode when cell-specific marker genes exist, which do not have to be known a priori, for each of constituent cell types. We demonstrate the performance of unsupervised deconvolution on both simulation and real gene expression data, together with perspective discussions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge