Rime Raissouni
Neural Multi-Hop Reasoning With Logical Rules on Biomedical Knowledge Graphs
Mar 18, 2021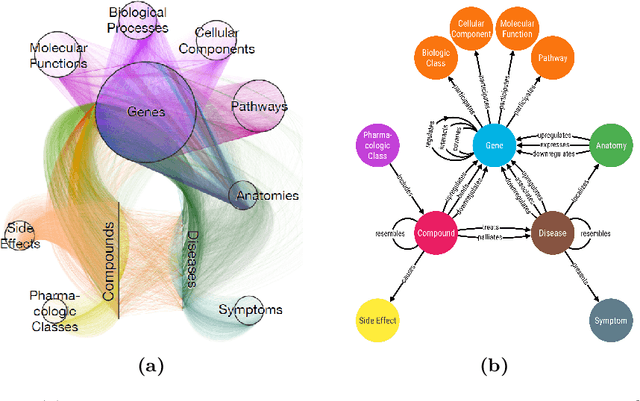

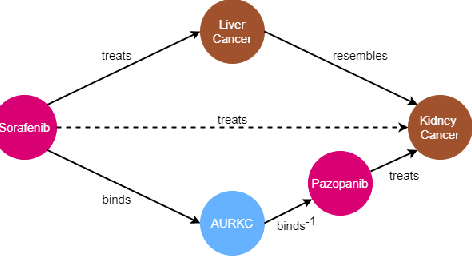
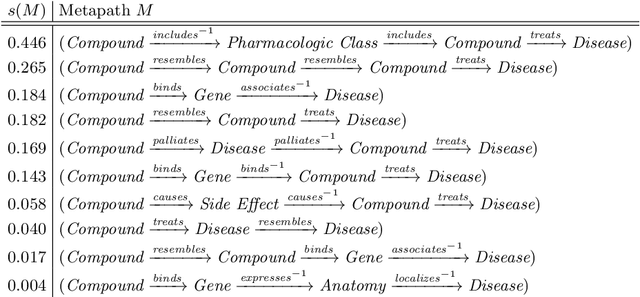
Abstract:Biomedical knowledge graphs permit an integrative computational approach to reasoning about biological systems. The nature of biological data leads to a graph structure that differs from those typically encountered in benchmarking datasets. To understand the implications this may have on the performance of reasoning algorithms, we conduct an empirical study based on the real-world task of drug repurposing. We formulate this task as a link prediction problem where both compounds and diseases correspond to entities in a knowledge graph. To overcome apparent weaknesses of existing algorithms, we propose a new method, PoLo, that combines policy-guided walks based on reinforcement learning with logical rules. These rules are integrated into the algorithm by using a novel reward function. We apply our method to Hetionet, which integrates biomedical information from 29 prominent bioinformatics databases. Our experiments show that our approach outperforms several state-of-the-art methods for link prediction while providing interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge