Ramy Arnaout
Which Similarity-Sensitive Entropy?
Nov 10, 2025Abstract:A canonical step in quantifying a system is to measure its entropy. Shannon entropy and other traditional entropy measures capture only the information encoded in the frequencies of a system's elements. Recently, Leinster, Cobbold, and Reeve (LCR) introduced a method that also captures the rich information encoded in the similarities and differences among elements, yielding similarity-sensitive entropy. More recently, the Vendi score (VS) was introduced as an alternative, raising the question of how LCR and VS compare, and which is preferable. Here we address these questions conceptually, analytically, and experimentally, using 53 machine-learning datasets. We show that LCR and VS can differ by orders of magnitude and can capture complementary information about a system, except in limiting cases. We demonstrate that both LCR and VS depend on how similarities are scaled and introduce the concept of ``half distance'' to parameterize this dependence. We prove that VS provides an upper bound on LCR for several values of the Rényi-Hill order parameter and conjecture that this bound holds for all values. We conclude that VS is preferable only when interpreting elements as linear combinations of a more fundamental set of ``ur-elements'' or when the system or dataset possesses a quantum-mechanical character. In the broader circumstance where one seeks simply to capture the rich information encoded by similarity, LCR is favored; nevertheless, for certain half-distances the two methods can complement each other.
X-Factor: Quality Is a Dataset-Intrinsic Property
May 28, 2025Abstract:In the universal quest to optimize machine-learning classifiers, three factors -- model architecture, dataset size, and class balance -- have been shown to influence test-time performance but do not fully account for it. Previously, evidence was presented for an additional factor that can be referred to as dataset quality, but it was unclear whether this was actually a joint property of the dataset and the model architecture, or an intrinsic property of the dataset itself. If quality is truly dataset-intrinsic and independent of model architecture, dataset size, and class balance, then the same datasets should perform better (or worse) regardless of these other factors. To test this hypothesis, here we create thousands of datasets, each controlled for size and class balance, and use them to train classifiers with a wide range of architectures, from random forests and support-vector machines to deep networks. We find that classifier performance correlates strongly by subset across architectures ($R^2=0.79$), supporting quality as an intrinsic property of datasets independent of dataset size and class balance and of model architecture. Digging deeper, we find that dataset quality appears to be an emergent property of something more fundamental: the quality of datasets' constituent classes. Thus, quality joins size, class balance, and model architecture as an independent correlate of performance and a separate target for optimizing machine-learning-based classification.
Grade Inflation in Generative Models
Jan 05, 2025Abstract:Generative models hold great potential, but only if one can trust the evaluation of the data they generate. We show that many commonly used quality scores for comparing two-dimensional distributions of synthetic vs. ground-truth data give better results than they should, a phenomenon we call the "grade inflation problem." We show that the correlation score, Jaccard score, earth-mover's score, and Kullback-Leibler (relative-entropy) score all suffer grade inflation. We propose that any score that values all datapoints equally, as these do, will also exhibit grade inflation; we refer to such scores as "equipoint" scores. We introduce the concept of "equidensity" scores, and present the Eden score, to our knowledge the first example of such a score. We found that Eden avoids grade inflation and agrees better with human perception of goodness-of-fit than the equipoint scores above. We propose that any reasonable equidensity score will avoid grade inflation. We identify a connection between equidensity scores and R\'enyi entropy of negative order. We conclude that equidensity scores are likely to outperform equipoint scores for generative models, and for comparing low-dimensional distributions more generally.
$\textit{lucie}$: An Improved Python Package for Loading Datasets from the UCI Machine Learning Repository
Oct 10, 2024Abstract:The University of California--Irvine (UCI) Machine Learning (ML) Repository (UCIMLR) is consistently cited as one of the most popular dataset repositories, hosting hundreds of high-impact datasets. However, a significant portion, including 28.4% of the top 250, cannot be imported via the $\textit{ucimlrepo}$ package that is provided and recommended by the UCIMLR website. Instead, they are hosted as .zip files, containing nonstandard formats that are difficult to import without additional ad hoc processing. To address this issue, here we present $\textit{lucie}$ -- $\underline{l}oad$ $\underline{U}niversity$ $\underline{C}alifornia$ $\underline{I}rvine$ $\underline{e}xamples$ -- a utility that automatically determines the data format and imports many of these previously non-importable datasets, while preserving as much of a tabular data structure as possible. $\textit{lucie}$ was designed using the top 100 most popular datasets and benchmarked on the next 130, where it resulted in a success rate of 95.4% vs. 73.1% for $\textit{ucimlrepo}$. $\textit{lucie}$ is available as a Python package on PyPI with 98% code coverage.
Beyond Size and Class Balance: Alpha as a New Dataset Quality Metric for Deep Learning
Jul 22, 2024Abstract:In deep learning, achieving high performance on image classification tasks requires diverse training sets. However, dataset diversity is incompletely understood. The current best practice is to try to maximize dataset size and class balance. Yet large, class-balanced datasets are not guaranteed to be diverse: images can still be arbitrarily similar. We hypothesized that, for a given model architecture, better model performance can be achieved by maximizing dataset diversity more directly. This could open a path for performance improvement without additional computational resources or architectural advances. To test this hypothesis, we introduce a comprehensive framework of diversity measures, developed in ecology, that generalizes familiar quantities like Shannon entropy by accounting for similarities and differences among images. (Dataset size and class balance emerge from this framework as special cases.) By analyzing thousands of subsets from seven medical datasets representing ultrasound, X-ray, CT, and pathology images, we found that the best correlates of performance were not size or class balance but $A$ -- ``big alpha'' -- a set of generalized entropy measures interpreted as the effective number of image-class pairs in the dataset, after accounting for similarities among images. One of these, $A_0$, explained 67\% of the variance in balanced accuracy across all subsets, vs. 54\% for class balance and just 39\% for size. The best pair was size and $A_1$ (79\%), which outperformed size and class balance (74\%). $A$ performed best for subsets from individual datasets as well as across datasets, supporting the generality of these results. We propose maximizing $A$ as a potential new way to improve the performance of deep learning in medical imaging.
Fast and accurate classification of echocardiograms using deep learning
Jun 27, 2017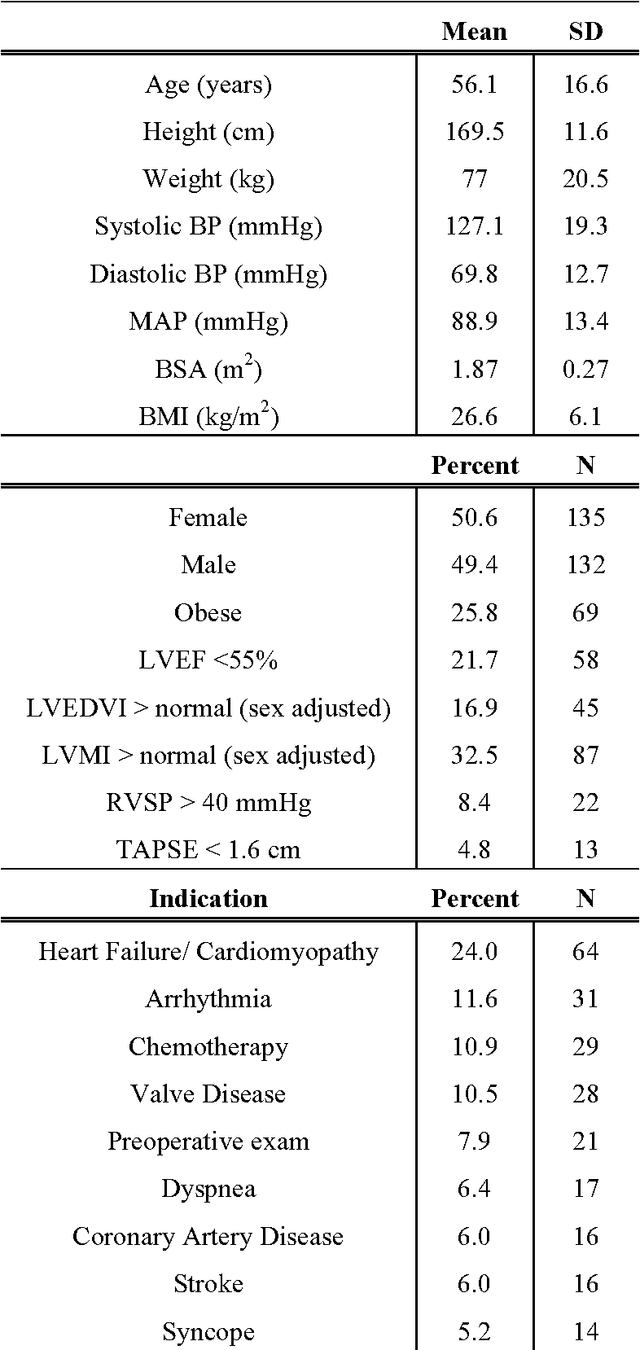
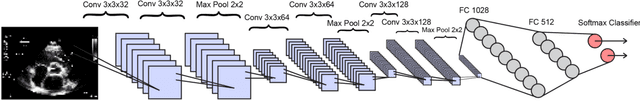
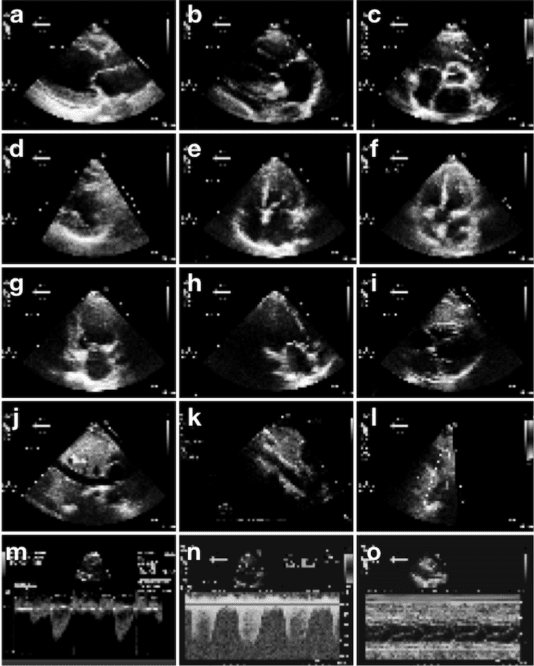
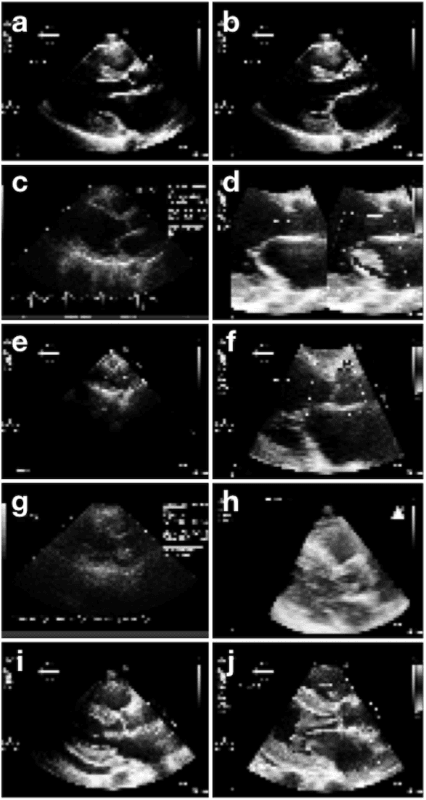
Abstract:Echocardiography is essential to modern cardiology. However, human interpretation limits high throughput analysis, limiting echocardiography from reaching its full clinical and research potential for precision medicine. Deep learning is a cutting-edge machine-learning technique that has been useful in analyzing medical images but has not yet been widely applied to echocardiography, partly due to the complexity of echocardiograms' multi view, multi modality format. The essential first step toward comprehensive computer assisted echocardiographic interpretation is determining whether computers can learn to recognize standard views. To this end, we anonymized 834,267 transthoracic echocardiogram (TTE) images from 267 patients (20 to 96 years, 51 percent female, 26 percent obese) seen between 2000 and 2017 and labeled them according to standard views. Images covered a range of real world clinical variation. We built a multilayer convolutional neural network and used supervised learning to simultaneously classify 15 standard views. Eighty percent of data used was randomly chosen for training and 20 percent reserved for validation and testing on never seen echocardiograms. Using multiple images from each clip, the model classified among 12 video views with 97.8 percent overall test accuracy without overfitting. Even on single low resolution images, test accuracy among 15 views was 91.7 percent versus 70.2 to 83.5 percent for board-certified echocardiographers. Confusional matrices, occlusion experiments, and saliency mapping showed that the model finds recognizable similarities among related views and classifies using clinically relevant image features. In conclusion, deep neural networks can classify essential echocardiographic views simultaneously and with high accuracy. Our results provide a foundation for more complex deep learning assisted echocardiographic interpretation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge