Rajat Mani Thomas
fMRI-S4: learning short- and long-range dynamic fMRI dependencies using 1D Convolutions and State Space Models
Aug 08, 2022


Abstract:Single-subject mapping of resting-state brain functional activity to non-imaging phenotypes is a major goal of neuroimaging. The large majority of learning approaches applied today rely either on static representations or on short-term temporal correlations. This is at odds with the nature of brain activity which is dynamic and exhibit both short- and long-range dependencies. Further, new sophisticated deep learning approaches have been developed and validated on single tasks/datasets. The application of these models for the study of a different targets typically require exhaustive hyperparameter search, model engineering and trial and error to obtain competitive results with simpler linear models. This in turn limit their adoption and hinder fair benchmarking in a rapidly developing area of research. To this end, we propose fMRI-S4; a versatile deep learning model for the classification of phenotypes and psychiatric disorders from the timecourses of resting-state functional magnetic resonance imaging scans. fMRI-S4 capture short- and long- range temporal dependencies in the signal using 1D convolutions and the recently introduced state-space models S4. The proposed architecture is lightweight, sample-efficient and robust across tasks/datasets. We validate fMRI-S4 on the tasks of diagnosing major depressive disorder (MDD), autism spectrum disorder (ASD) and sex classifcation on three multi-site rs-fMRI datasets. We show that fMRI-S4 can outperform existing methods on all three tasks and can be trained as a plug&play model without special hyperpararameter tuning for each setting
Improving the Diagnosis of Psychiatric Disorders with Self-Supervised Graph State Space Models
Jun 07, 2022



Abstract:Single subject prediction of brain disorders from neuroimaging data has gained increasing attention in recent years. Yet, for some heterogeneous disorders such as major depression disorder (MDD) and autism spectrum disorder (ASD), the performance of prediction models on large-scale multi-site datasets remains poor. We present a two-stage framework to improve the diagnosis of heterogeneous psychiatric disorders from resting-state functional magnetic resonance imaging (rs-fMRI). First, we propose a self-supervised mask prediction task on data from healthy individuals that can exploit differences between healthy controls and patients in clinical datasets. Next, we train a supervised classifier on the learned discriminative representations. To model rs-fMRI data, we develop Graph-S4; an extension to the recently proposed state-space model S4 to graph settings where the underlying graph structure is not known in advance. We show that combining the framework and Graph-S4 can significantly improve the diagnostic performance of neuroimaging-based single subject prediction models of MDD and ASD on three open-source multi-center rs-fMRI clinical datasets.
Dynamic Adaptive Spatio-temporal Graph Convolution for fMRI Modelling
Sep 26, 2021



Abstract:The characterisation of the brain as a functional network in which the connections between brain regions are represented by correlation values across time series has been very popular in the last years. Although this representation has advanced our understanding of brain function, it represents a simplified model of brain connectivity that has a complex dynamic spatio-temporal nature. Oversimplification of the data may hinder the merits of applying advanced non-linear feature extraction algorithms. To this end, we propose a dynamic adaptive spatio-temporal graph convolution (DAST-GCN) model to overcome the shortcomings of pre-defined static correlation-based graph structures. The proposed approach allows end-to-end inference of dynamic connections between brain regions via layer-wise graph structure learning module while mapping brain connectivity to a phenotype in a supervised learning framework. This leverages the computational power of the model, data and targets to represent brain connectivity, and could enable the identification of potential biomarkers for the supervised target in question. We evaluate our pipeline on the UKBiobank dataset for age and gender classification tasks from resting-state functional scans and show that it outperforms currently adapted linear and non-linear methods in neuroimaging. Further, we assess the generalizability of the inferred graph structure by transferring the pre-trained graph to an independent dataset for the same task. Our results demonstrate the task-robustness of the graph against different scanning parameters and demographics.
* Accepted at International Workshop on Machine Learning in Clinical Neuroimaging (MLCN2021)
Improving Solar Cell Metallization Designs using Convolutional Neural Networks
Apr 08, 2021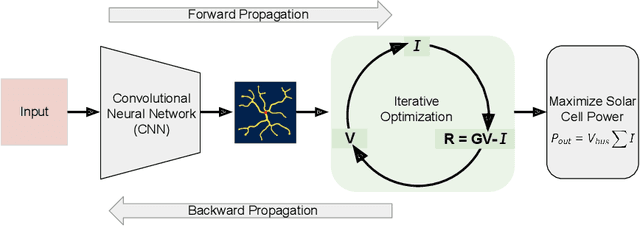
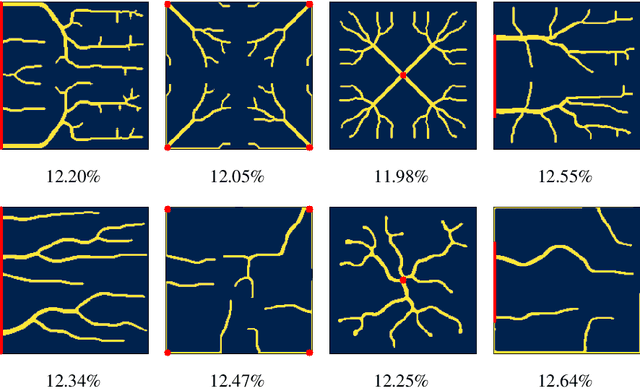
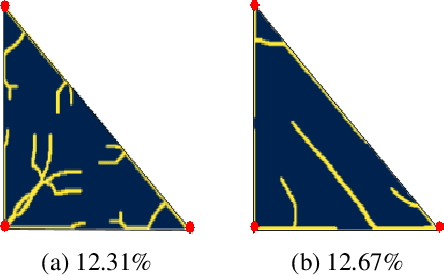
Abstract:Optimizing the design of solar cell metallizations is one of the ways to improve the performance of solar cells. Recently, it has been shown that Topology Optimization (TO) can be used to design complex metallization patterns for solar cells that lead to improved performance. TO generates unconventional design patterns that cannot be obtained with the traditional shape optimization methods. In this paper, we show that this design process can be improved further using a deep learning inspired strategy. We present SolarNet, a CNN-based reparameterization scheme that can be used to obtain improved metallization designs. SolarNet modifies the optimization domain such that rather than optimizing the electrode material distribution directly, the weights of a CNN model are optimized. The design generated by CNN is then evaluated using the physics equations, which in turn generates gradients for backpropagation. SolarNet is trained end-to-end involving backpropagation through the solar cell model as well as the CNN pipeline. Through application on solar cells of different shapes as well as different busbar geometries, we demonstrate that SolarNet improves the performance of solar cells compared to the traditional TO approach.
A Hybrid 3DCNN and 3DC-LSTM based model for 4D Spatio-temporal fMRI data: An ABIDE Autism Classification study
Feb 14, 2020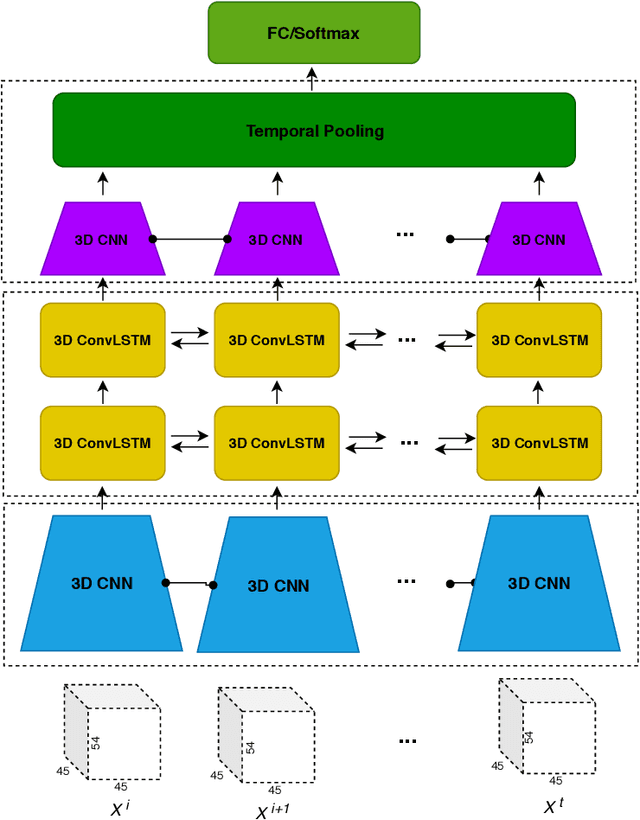
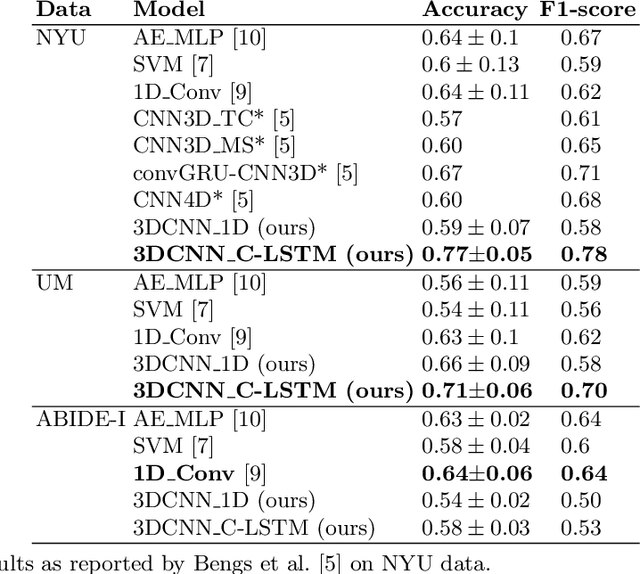
Abstract:Functional Magnetic Resonance Imaging (fMRI) captures the temporal dynamics of neural activity as a function of spatial location in the brain. Thus, fMRI scans are represented as 4-Dimensional (3-space + 1-time) tensors. And it is widely believed that the spatio-temporal patterns in fMRI manifests as behaviour and clinical symptoms. Because of the high dimensionality ($\sim$ 1 Million) of fMRI, and the added constraints of limited cardinality of data sets, extracting such patterns are challenging. A standard approach to overcome these hurdles is to reduce the dimensionality of the data by either summarizing activation over time or space at the expense of possible loss of useful information. Here, we introduce an end-to-end algorithm capable of extracting spatiotemporal features from the full 4-D data using 3-D CNNs and 3-D Convolutional LSTMs. We evaluate our proposed model on the publicly available ABIDE dataset to demonstrate the capability of our model to classify Autism Spectrum Disorder (ASD) from resting-state fMRI data. Our results show that the proposed model achieves state of the art results on single sites with F1-scores of 0.78 and 0.7 on NYU and UM sites, respectively.
* 8pages
Simple 1-D Convolutional Networks for Resting-State fMRI Based Classification in Autism
Jul 02, 2019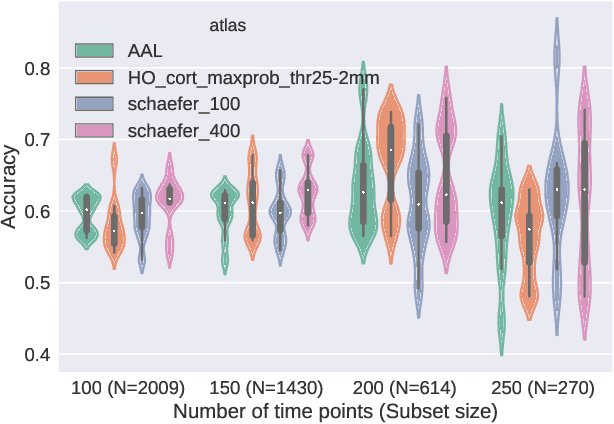
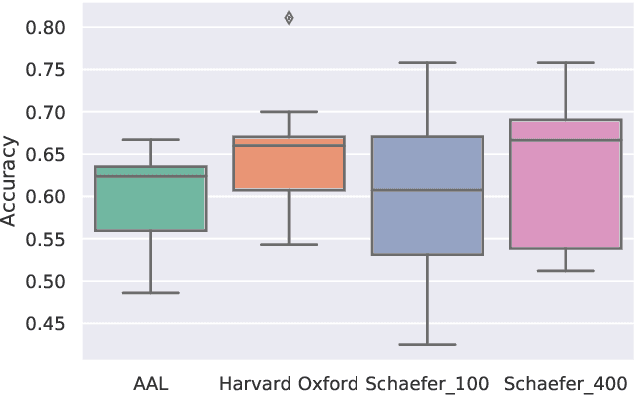
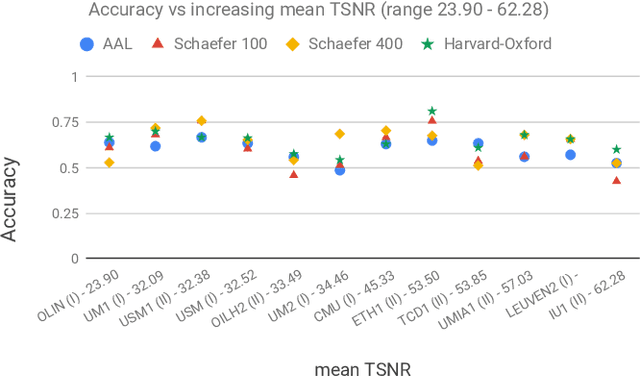
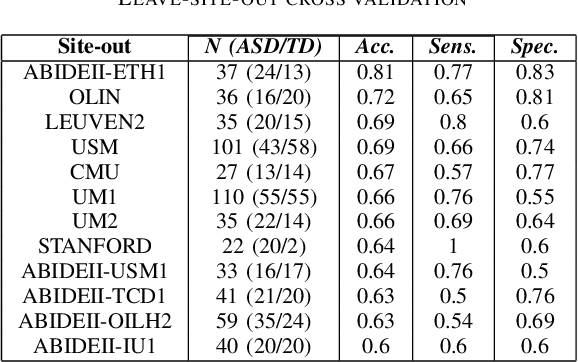
Abstract:Deep learning methods are increasingly being used with neuroimaging data like structural and function magnetic resonance imaging (MRI) to predict the diagnosis of neuropsychiatric and neurological disorders. For psychiatric disorders in particular, it is believed that one of the most promising modality is the resting-state functional MRI (rsfMRI), which captures the intrinsic connectivity between regions in the brain. Because rsfMRI data points are inherently high-dimensional (~1M), it is impossible to process the entire input in its raw form. In this paper, we propose a very simple transformation of the rsfMRI images that captures all of the temporal dynamics of the signal but sub-samples its spatial extent. As a result, we use a very simple 1-D convolutional network which is fast to train, requires minimal preprocessing and performs at par with the state-of-the-art on the classification of Autism spectrum disorders.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge