Rahul Kukreja
Node Injection for Class-specific Network Poisoning
Jan 28, 2023Abstract:Graph Neural Networks (GNNs) are powerful in learning rich network representations that aid the performance of downstream tasks. However, recent studies showed that GNNs are vulnerable to adversarial attacks involving node injection and network perturbation. Among these, node injection attacks are more practical as they don't require manipulation in the existing network and can be performed more realistically. In this paper, we propose a novel problem statement - a class-specific poison attack on graphs in which the attacker aims to misclassify specific nodes in the target class into a different class using node injection. Additionally, nodes are injected in such a way that they camouflage as benign nodes. We propose NICKI, a novel attacking strategy that utilizes an optimization-based approach to sabotage the performance of GNN-based node classifiers. NICKI works in two phases - it first learns the node representation and then generates the features and edges of the injected nodes. Extensive experiments and ablation studies on four benchmark networks show that NICKI is consistently better than four baseline attacking strategies for misclassifying nodes in the target class. We also show that the injected nodes are properly camouflaged as benign, thus making the poisoned graph indistinguishable from its clean version w.r.t various topological properties.
DAGSurv: Directed Acyclic Graph Based Survival Analysis Using Deep Neural Networks
Nov 02, 2021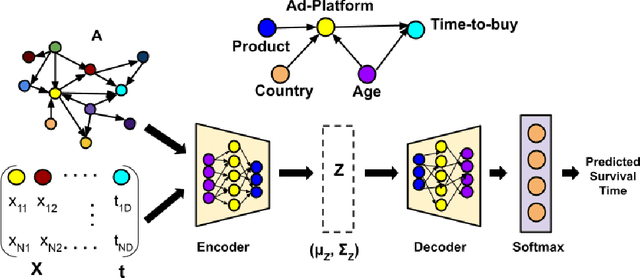
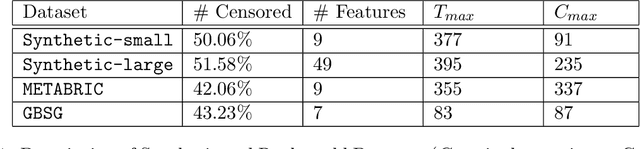
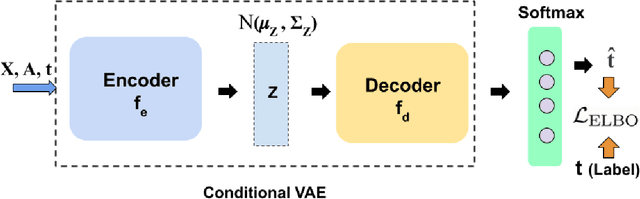
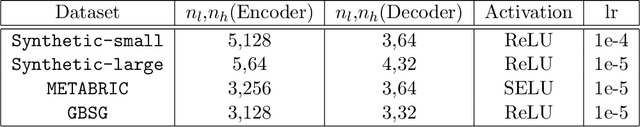
Abstract:Causal structures for observational survival data provide crucial information regarding the relationships between covariates and time-to-event. We derive motivation from the information theoretic source coding argument, and show that incorporating the knowledge of the directed acyclic graph (DAG) can be beneficial if suitable source encoders are employed. As a possible source encoder in this context, we derive a variational inference based conditional variational autoencoder for causal structured survival prediction, which we refer to as DAGSurv. We illustrate the performance of DAGSurv on low and high-dimensional synthetic datasets, and real-world datasets such as METABRIC and GBSG. We demonstrate that the proposed method outperforms other survival analysis baselines such as Cox Proportional Hazards, DeepSurv and Deephit, which are oblivious to the underlying causal relationship between data entities.
BrainFrame: A node-level heterogeneous accelerator platform for neuron simulations
Aug 15, 2017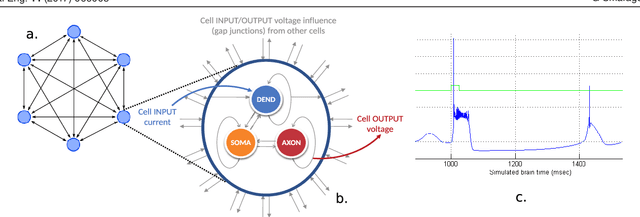

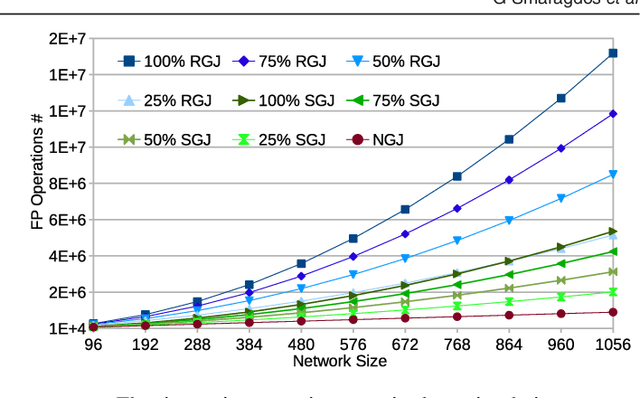

Abstract:Objective: The advent of High-Performance Computing (HPC) in recent years has led to its increasing use in brain study through computational models. The scale and complexity of such models are constantly increasing, leading to challenging computational requirements. Even though modern HPC platforms can often deal with such challenges, the vast diversity of the modeling field does not permit for a single acceleration (or homogeneous) platform to effectively address the complete array of modeling requirements. Approach: In this paper we propose and build BrainFrame, a heterogeneous acceleration platform, incorporating three distinct acceleration technologies, a Dataflow Engine, a Xeon Phi and a GP-GPU. The PyNN framework is also integrated into the platform. As a challenging proof of concept, we analyze the performance of BrainFrame on different instances of a state-of-the-art neuron model, modeling the Inferior- Olivary Nucleus using a biophysically-meaningful, extended Hodgkin-Huxley representation. The model instances take into account not only the neuronal- network dimensions but also different network-connectivity circumstances that can drastically change application workload characteristics. Main results: The synthetic approach of three HPC technologies demonstrated that BrainFrame is better able to cope with the modeling diversity encountered. Our performance analysis shows clearly that the model directly affect performance and all three technologies are required to cope with all the model use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge