Peikun Guo
ECG Semantic Integrator (ESI): A Foundation ECG Model Pretrained with LLM-Enhanced Cardiological Text
May 26, 2024



Abstract:The utilization of deep learning on electrocardiogram (ECG) analysis has brought the advanced accuracy and efficiency of cardiac healthcare diagnostics. By leveraging the capabilities of deep learning in semantic understanding, especially in feature extraction and representation learning, this study introduces a new multimodal contrastive pretaining framework that aims to improve the quality and robustness of learned representations of 12-lead ECG signals. Our framework comprises two key components, including Cardio Query Assistant (CQA) and ECG Semantics Integrator(ESI). CQA integrates a retrieval-augmented generation (RAG) pipeline to leverage large language models (LLMs) and external medical knowledge to generate detailed textual descriptions of ECGs. The generated text is enriched with information about demographics and waveform patterns. ESI integrates both contrastive and captioning loss to pretrain ECG encoders for enhanced representations. We validate our approach through various downstream tasks, including arrhythmia detection and ECG-based subject identification. Our experimental results demonstrate substantial improvements over strong baselines in these tasks. These baselines encompass supervised and self-supervised learning methods, as well as prior multimodal pretraining approaches.
AdaWaveNet: Adaptive Wavelet Network for Time Series Analysis
May 17, 2024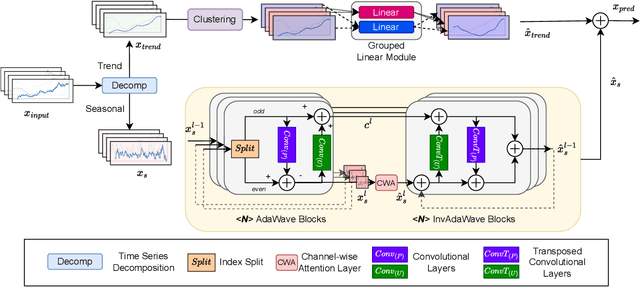
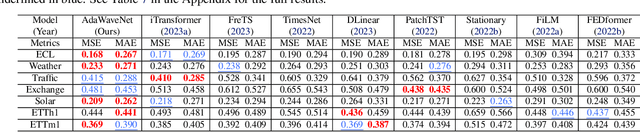
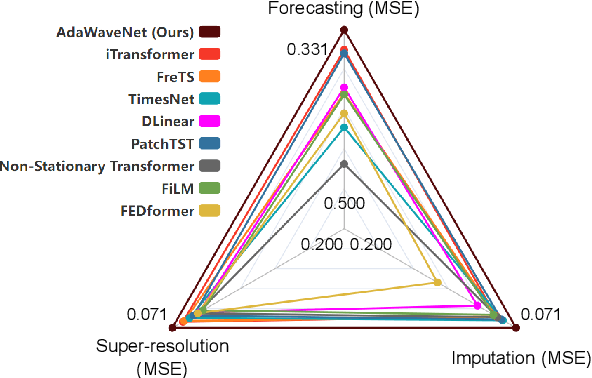
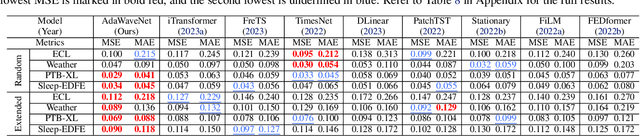
Abstract:Time series data analysis is a critical component in various domains such as finance, healthcare, and meteorology. Despite the progress in deep learning for time series analysis, there remains a challenge in addressing the non-stationary nature of time series data. Traditional models, which are built on the assumption of constant statistical properties over time, often struggle to capture the temporal dynamics in realistic time series, resulting in bias and error in time series analysis. This paper introduces the Adaptive Wavelet Network (AdaWaveNet), a novel approach that employs Adaptive Wavelet Transformation for multi-scale analysis of non-stationary time series data. AdaWaveNet designed a lifting scheme-based wavelet decomposition and construction mechanism for adaptive and learnable wavelet transforms, which offers enhanced flexibility and robustness in analysis. We conduct extensive experiments on 10 datasets across 3 different tasks, including forecasting, imputation, and a newly established super-resolution task. The evaluations demonstrate the effectiveness of AdaWaveNet over existing methods in all three tasks, which illustrates its potential in various real-world applications.
Balanced Mixed-Type Tabular Data Synthesis with Diffusion Models
Apr 12, 2024Abstract:Diffusion models have emerged as a robust framework for various generative tasks, such as image and audio synthesis, and have also demonstrated a remarkable ability to generate mixed-type tabular data comprising both continuous and discrete variables. However, current approaches to training diffusion models on mixed-type tabular data tend to inherit the imbalanced distributions of features present in the training dataset, which can result in biased sampling. In this research, we introduce a fair diffusion model designed to generate balanced data on sensitive attributes. We present empirical evidence demonstrating that our method effectively mitigates the class imbalance in training data while maintaining the quality of the generated samples. Furthermore, we provide evidence that our approach outperforms existing methods for synthesizing tabular data in terms of performance and fairness.
Empirical Study of Mix-based Data Augmentation Methods in Physiological Time Series Data
Sep 18, 2023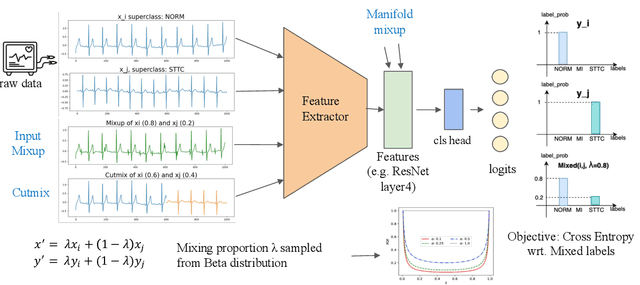
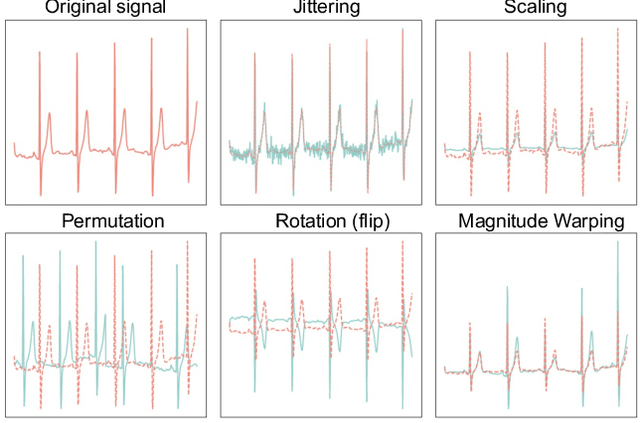
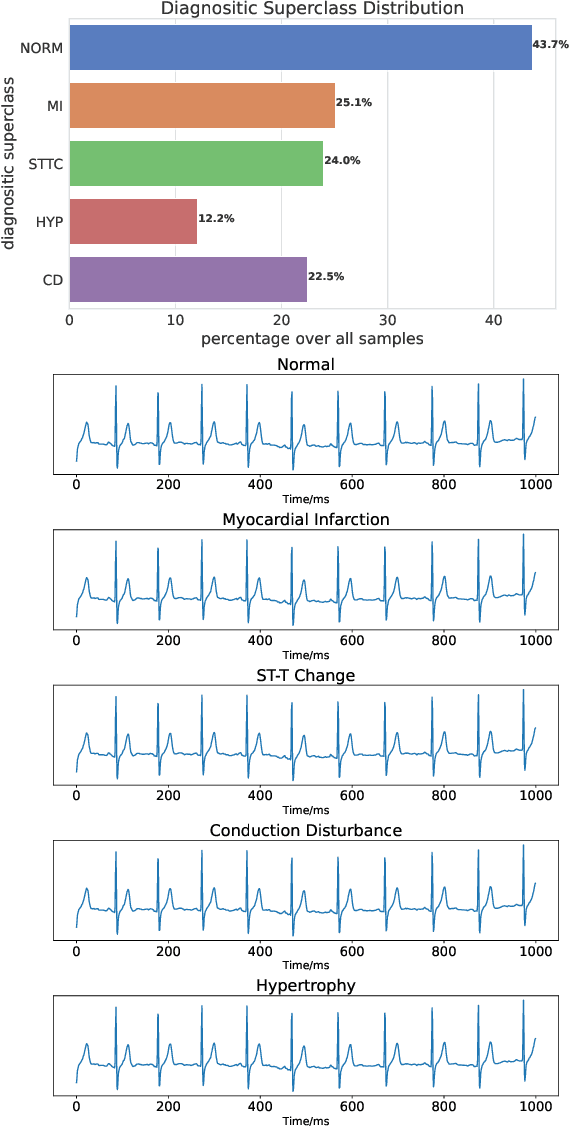
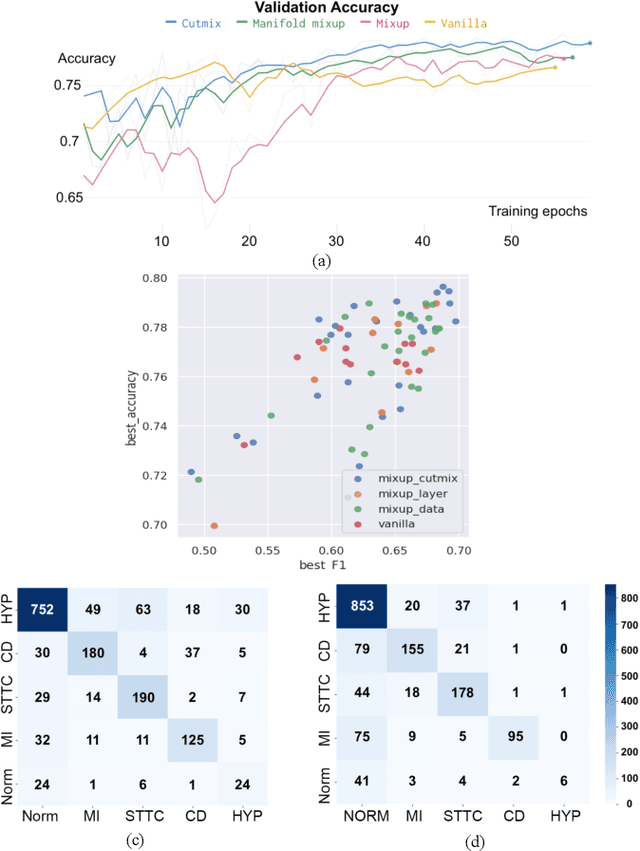
Abstract:Data augmentation is a common practice to help generalization in the procedure of deep model training. In the context of physiological time series classification, previous research has primarily focused on label-invariant data augmentation methods. However, another class of augmentation techniques (\textit{i.e., Mixup}) that emerged in the computer vision field has yet to be fully explored in the time series domain. In this study, we systematically review the mix-based augmentations, including mixup, cutmix, and manifold mixup, on six physiological datasets, evaluating their performance across different sensory data and classification tasks. Our results demonstrate that the three mix-based augmentations can consistently improve the performance on the six datasets. More importantly, the improvement does not rely on expert knowledge or extensive parameter tuning. Lastly, we provide an overview of the unique properties of the mix-based augmentation methods and highlight the potential benefits of using the mix-based augmentation in physiological time series data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge