Pedro Silva
Benford's Law as a Distributional Prior for Post-Training Quantization of Large Language Models
Jan 29, 2026Abstract:The rapid growth of Large Language Models (LLMs) intensifies the need for effective compression, with weight quantization being the most widely adopted technique. Standard uniform quantizers assume that parameters are evenly distributed, an assumption at odds with the highly skewed distributions observed in practice. We propose Benford-Quant, a simple, data-free non-uniform quantizer inspired by Benford's Law, which predicts that leading digits follow a logarithmic distribution. Benford-Quant replaces the uniform grid with a log-spaced codebook, dedicating more resolution to the frequent small-magnitude weights. We provide both theoretical intuition and empirical evidence: (i) weights in transformer transformational layers adhere closely to Benford statistics, while normalization layers systematically deviate; (ii) on Small Language Models (SLMs), Benford-Quant consistently improves perplexity, reducing 4-bit perplexity on Gemma-270M by more than 10%; and (iii) on larger LLMs, it remains competitive, with differences explained by over-parameterization effects. Our results indicate that incorporating a Benford-inspired prior into quantization grids is a low-cost modification that yields accuracy gains in aggressive few-bit regimes. Although it is not able to surpass the state of the art in tasks such as perplexity and LAMBADA, the Benford-Quant approach can be hybridized with other quantization methods-such as SmoothQuant and Activation-Aware Quantization-without major pipeline modification, potentially improving their performance.
PD-Loss: Proxy-Decidability for Efficient Metric Learning
Aug 23, 2025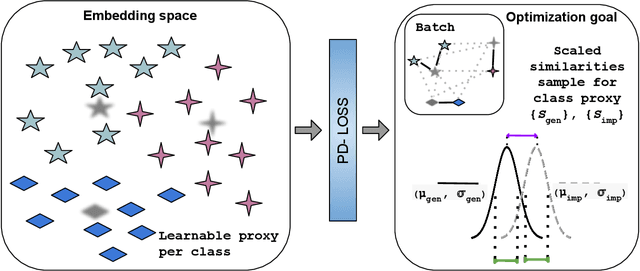
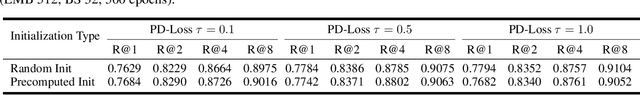
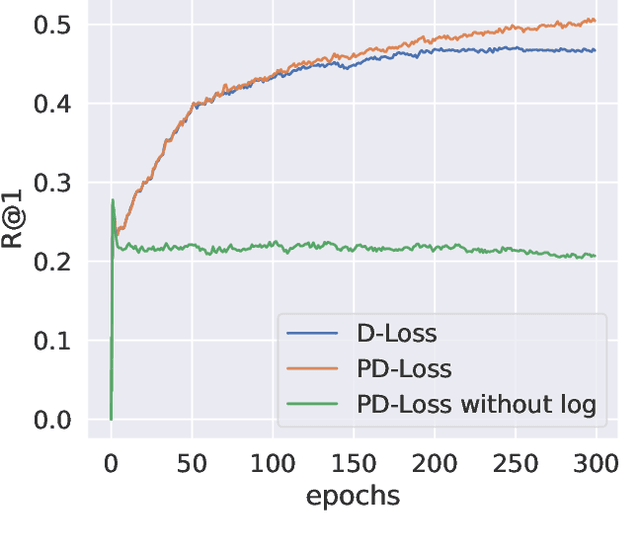
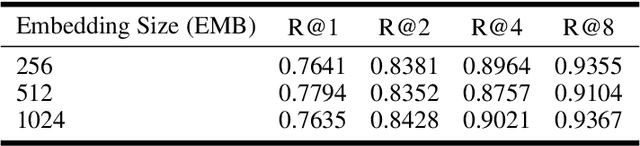
Abstract:Deep Metric Learning (DML) aims to learn embedding functions that map semantically similar inputs to proximate points in a metric space while separating dissimilar ones. Existing methods, such as pairwise losses, are hindered by complex sampling requirements and slow convergence. In contrast, proxy-based losses, despite their improved scalability, often fail to optimize global distribution properties. The Decidability-based Loss (D-Loss) addresses this by targeting the decidability index (d') to enhance distribution separability, but its reliance on large mini-batches imposes significant computational constraints. We introduce Proxy-Decidability Loss (PD-Loss), a novel objective that integrates learnable proxies with the statistical framework of d' to optimize embedding spaces efficiently. By estimating genuine and impostor distributions through proxies, PD-Loss combines the computational efficiency of proxy-based methods with the principled separability of D-Loss, offering a scalable approach to distribution-aware DML. Experiments across various tasks, including fine-grained classification and face verification, demonstrate that PD-Loss achieves performance comparable to that of state-of-the-art methods while introducing a new perspective on embedding optimization, with potential for broader applications.
A Systematic Review of ECG Arrhythmia Classification: Adherence to Standards, Fair Evaluation, and Embedded Feasibility
Mar 10, 2025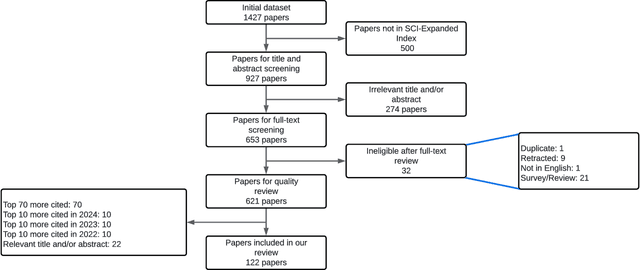
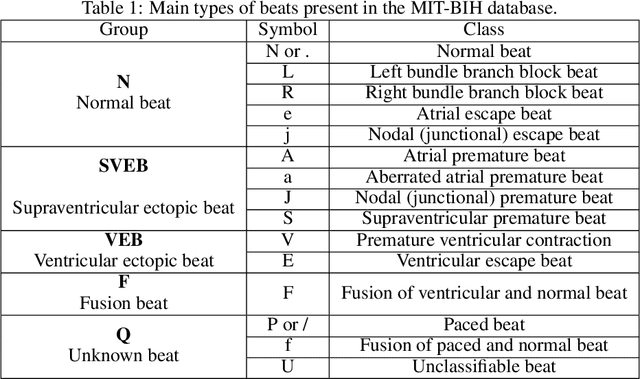
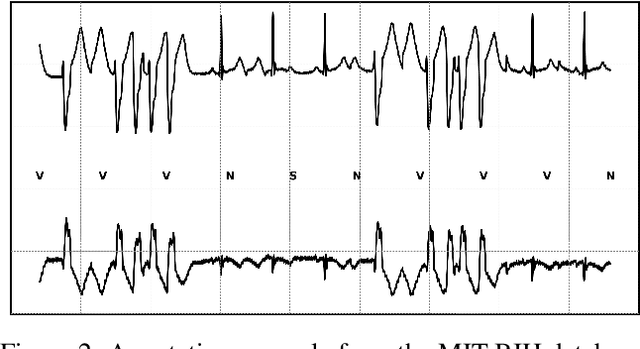
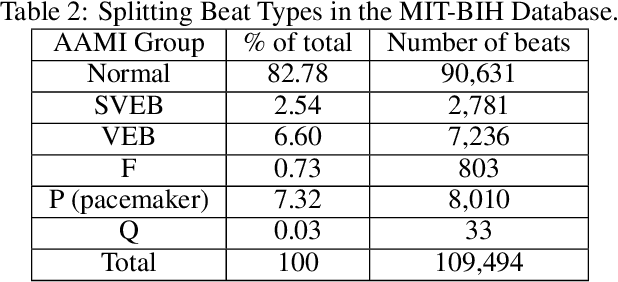
Abstract:The classification of electrocardiogram (ECG) signals is crucial for early detection of arrhythmias and other cardiac conditions. However, despite advances in machine learning, many studies fail to follow standardization protocols, leading to inconsistencies in performance evaluation and real-world applicability. Additionally, hardware constraints essential for practical deployment, such as in pacemakers, Holter monitors, and wearable ECG patches, are often overlooked. Since real-world impact depends on feasibility in resource-constrained devices, ensuring efficient deployment is critical for continuous monitoring. This review systematically analyzes ECG classification studies published between 2017 and 2024, focusing on those adhering to the E3C (Embedded, Clinical, and Comparative Criteria), which include inter-patient paradigm implementation, compliance with Association for the Advancement of Medical Instrumentation (AAMI) recommendations, and model feasibility for embedded systems. While many studies report high accuracy, few properly consider patient-independent partitioning and hardware limitations. We identify state-of-the-art methods meeting E3C criteria and conduct a comparative analysis of accuracy, inference time, energy consumption, and memory usage. Finally, we propose standardized reporting practices to ensure fair comparisons and practical applicability of ECG classification models. By addressing these gaps, this study aims to guide future research toward more robust and clinically viable ECG classification systems.
Representation Online Matters: Practical End-to-End Diversification in Search and Recommender Systems
May 26, 2023Abstract:As the use of online platforms continues to grow across all demographics, users often express a desire to feel represented in the content. To improve representation in search results and recommendations, we introduce end-to-end diversification, ensuring that diverse content flows throughout the various stages of these systems, from retrieval to ranking. We develop, experiment, and deploy scalable diversification mechanisms in multiple production surfaces on the Pinterest platform, including Search, Related Products, and New User Homefeed, to improve the representation of different skin tones in beauty and fashion content. Diversification in production systems includes three components: identifying requests that will trigger diversification, ensuring diverse content is retrieved from the large content corpus during the retrieval stage, and finally, balancing the diversity-utility trade-off in a self-adjusting manner in the ranking stage. Our approaches, which evolved from using Strong-OR logical operator to bucketized retrieval at the retrieval stage and from greedy re-rankers to multi-objective optimization using determinantal point processes for the ranking stage, balances diversity and utility while enabling fast iterations and scalable expansion to diversification over multiple dimensions. Our experiments indicate that these approaches significantly improve diversity metrics, with a neutral to a positive impact on utility metrics and improved user satisfaction, both qualitatively and quantitatively, in production. An accessible PDF of this article is available at https://drive.google.com/file/d/1p5PkqC-sdtX19Y_IAjZCtiSxSEX1IP3q/view
CapsProm: A Capsule Network For Promoter Prediction
Dec 07, 2021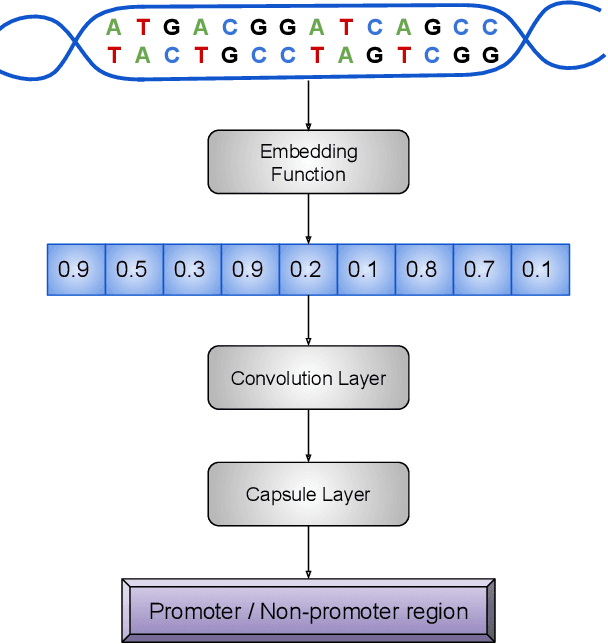
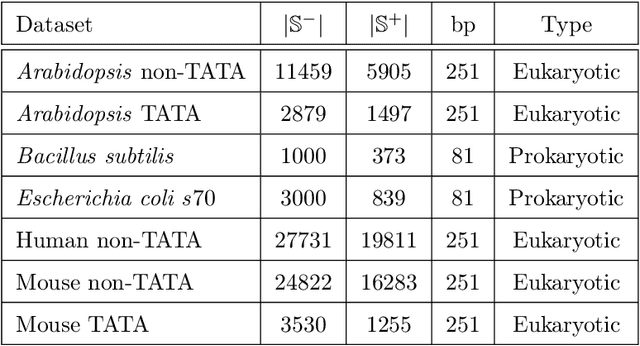
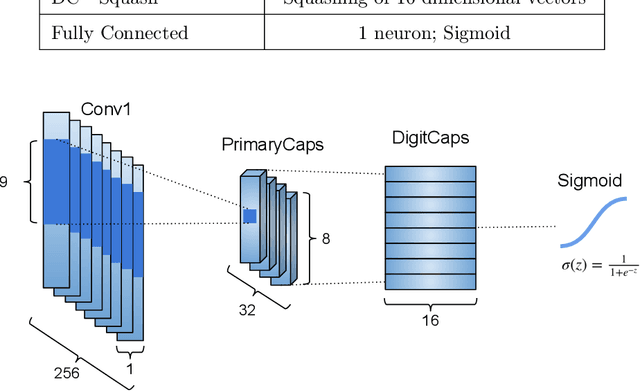
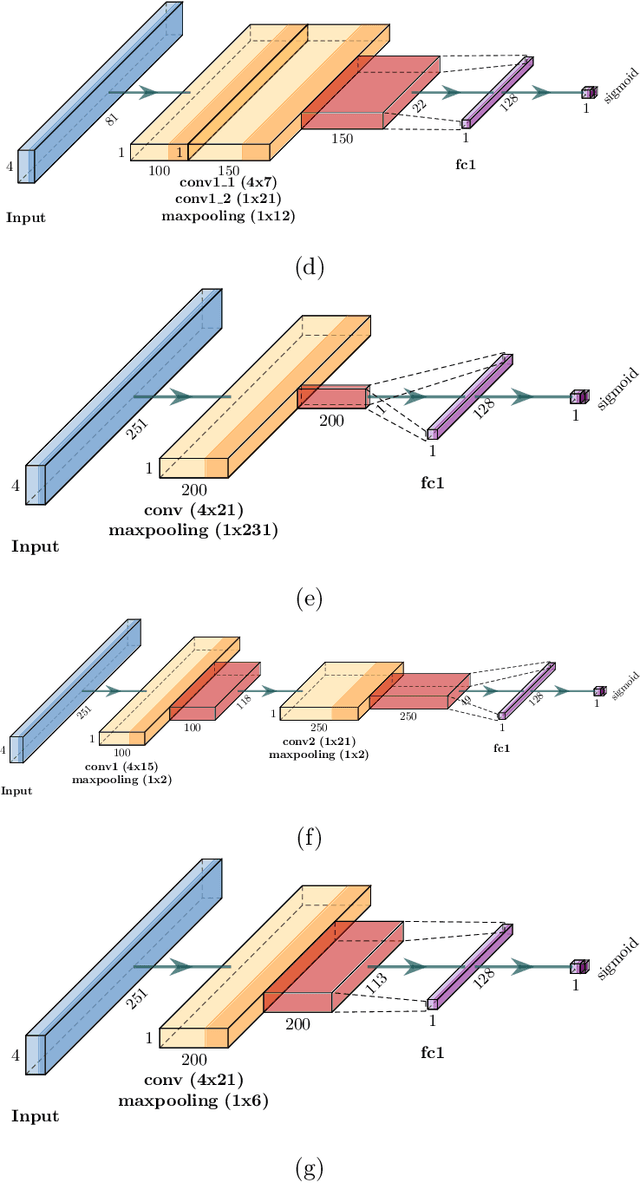
Abstract:Locating the promoter region in DNA sequences is of paramount importance in the field of bioinformatics. This is a problem widely studied in the literature, however, not yet fully resolved. Some researchers have presented remarkable results using convolution networks, that allowed the automatic extraction of features from a DNA chain. However, a universal architecture that could generalize to several organisms has not yet been achieved, and thus, requiring researchers to seek new architectures and hyperparameters for each new organism evaluated. In this work, we propose a versatile architecture, based on capsule network, that can accurately identify promoter sequences in raw DNA data from seven different organisms, eukaryotic, and prokaryotic. Our model, the CapsProm, could assist in the transfer of learning between organisms and expand its applicability. Furthermore the CapsProm showed competitive results, overcoming the baseline method in five out of seven of the tested datasets (F1-score). The models and source code are made available at https://github.com/lauromoraes/CapsNet-promoter.
A Decidability-Based Loss Function
Sep 12, 2021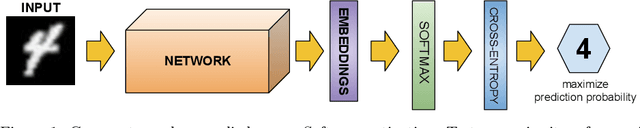
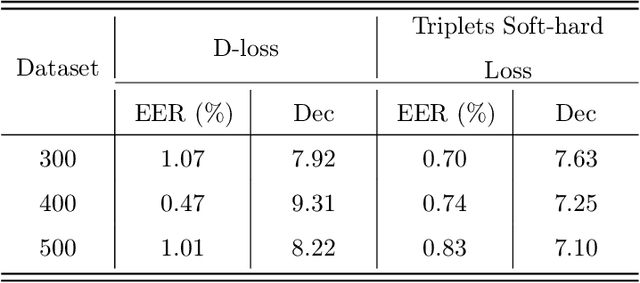
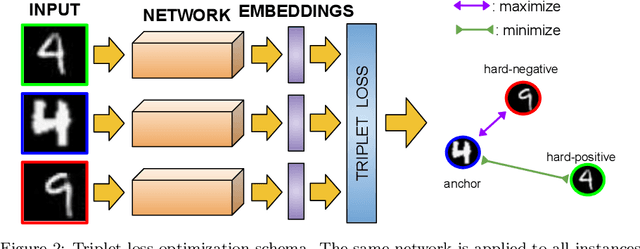
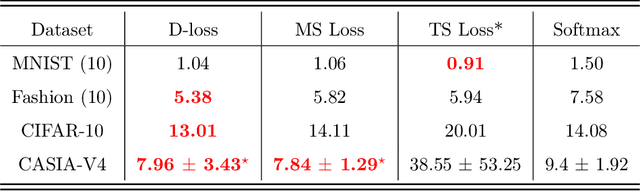
Abstract:Nowadays, deep learning is the standard approach for a wide range of problems, including biometrics, such as face recognition and speech recognition, etc. Biometric problems often use deep learning models to extract features from images, also known as embeddings. Moreover, the loss function used during training strongly influences the quality of the generated embeddings. In this work, a loss function based on the decidability index is proposed to improve the quality of embeddings for the verification routine. Our proposal, the D-loss, avoids some Triplet-based loss disadvantages such as the use of hard samples and tricky parameter tuning, which can lead to slow convergence. The proposed approach is compared against the Softmax (cross-entropy), Triplets Soft-Hard, and the Multi Similarity losses in four different benchmarks: MNIST, Fashion-MNIST, CIFAR10 and CASIA-IrisV4. The achieved results show the efficacy of the proposal when compared to other popular metrics in the literature. The D-loss computation, besides being simple, non-parametric and easy to implement, favors both the inter-class and intra-class scenarios.
Improving Malware Detection Accuracy by Extracting Icon Information
Dec 10, 2017
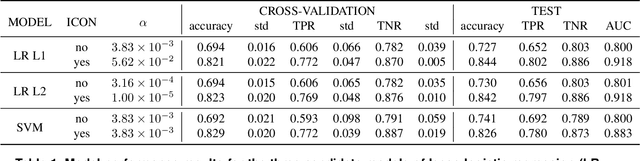
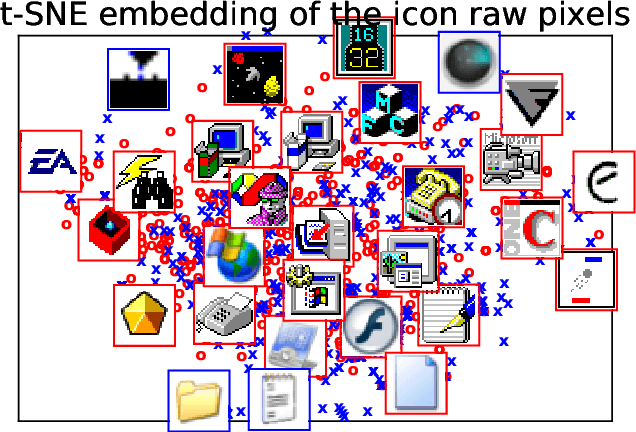
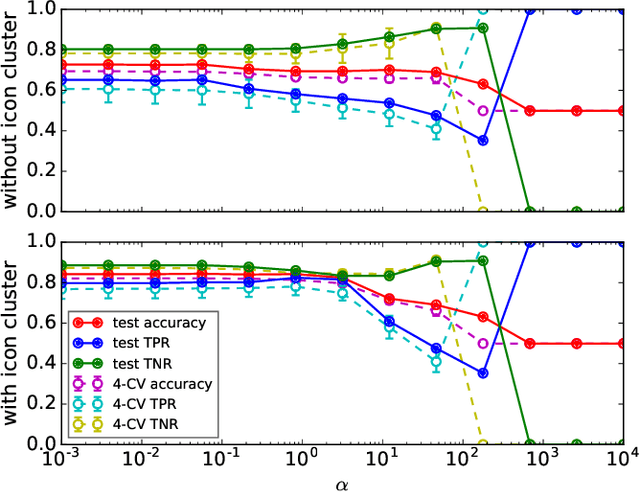
Abstract:Detecting PE malware files is now commonly approached using statistical and machine learning models. While these models commonly use features extracted from the structure of PE files, we propose that icons from these files can also help better predict malware. We propose an innovative machine learning approach to extract information from icons. Our proposed approach consists of two steps: 1) extracting icon features using summary statics, histogram of gradients (HOG), and a convolutional autoencoder, 2) clustering icons based on the extracted icon features. Using publicly available data and by using machine learning experiments, we show our proposed icon clusters significantly boost the efficacy of malware prediction models. In particular, our experiments show an average accuracy increase of 10% when icon clusters are used in the prediction model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge