Pablo Polosecki
GOKU-UI: Ubiquitous Inference through Attention and Multiple Shooting for Continuous-time Generative Models
Jul 11, 2023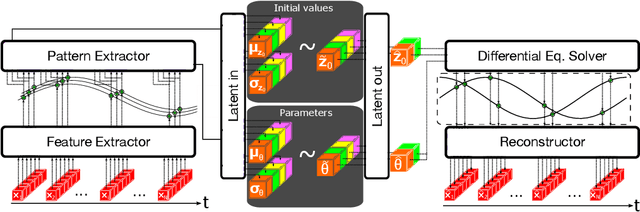
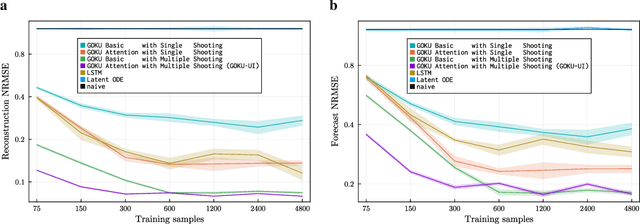
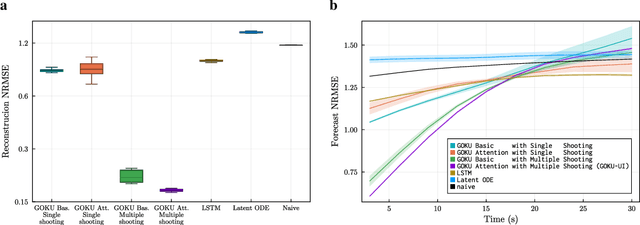

Abstract:Scientific Machine Learning (SciML) is a burgeoning field that synergistically combines domain-aware and interpretable models with agnostic machine learning techniques. In this work, we introduce GOKU-UI, an evolution of the SciML generative model GOKU-nets. The GOKU-UI broadens the original model's spectrum to incorporate other classes of differential equations, such as Stochastic Differential Equations (SDEs), and integrates a distributed, i.e. ubiquitous, inference through attention mechanisms and a novel multiple shooting training strategy in the latent space. These enhancements have led to a significant increase in its performance in both reconstruction and forecast tasks, as demonstrated by our evaluation of simulated and empirical data. Specifically, GOKU-UI outperformed all baseline models on synthetic datasets even with a training set 32-fold smaller, underscoring its remarkable data efficiency. Furthermore, when applied to empirical human brain data, while incorporating stochastic Stuart-Landau oscillators into its dynamical core, it not only surpassed state-of-the-art baseline methods in the reconstruction task, but also demonstrated better prediction of future brain activity up to 12 seconds ahead. By training GOKU-UI on resting-state fMRI data, we encoded whole-brain dynamics into a latent representation, learning an effective low-dimensional dynamical system model that could offer insights into brain functionality and open avenues for practical applications such as mental state or psychiatric condition classification. Ultimately, our research provides further impetus for the field of Scientific Machine Learning, showcasing the potential for advancements when established scientific insights are interwoven with modern machine learning.
Learning Nonlinear Brain Dynamics: van der Pol Meets LSTM
May 24, 2018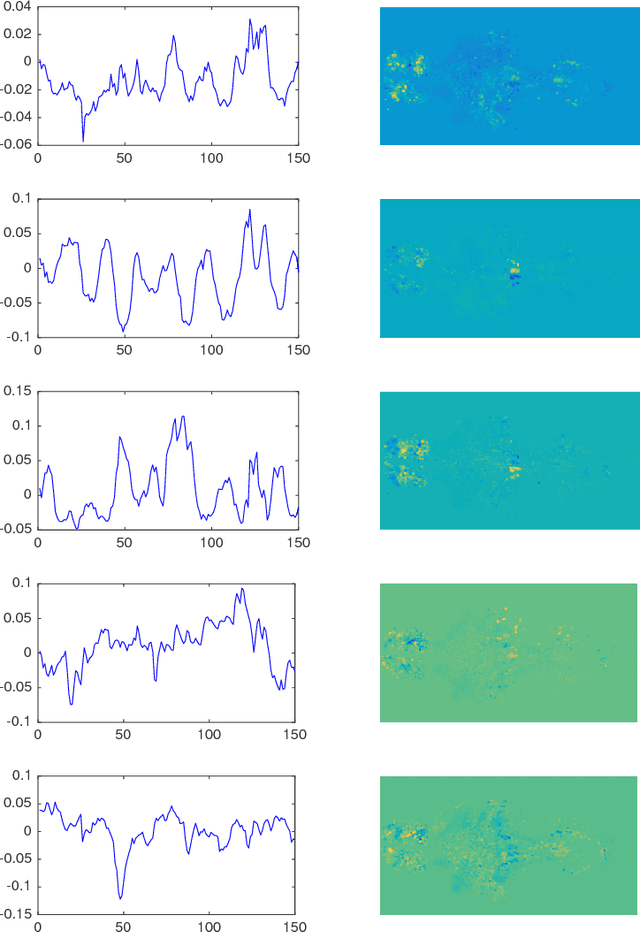
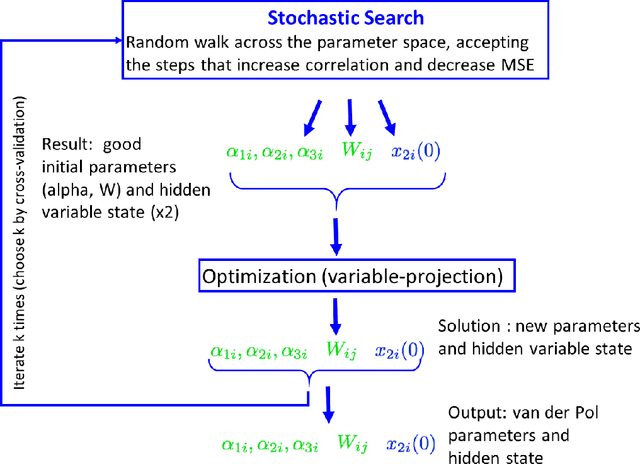
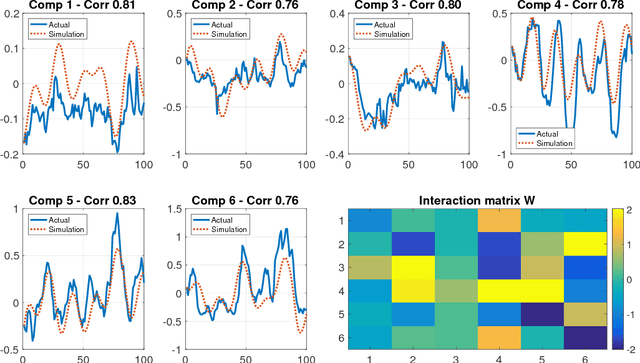
Abstract:Many real-world data sets, especially in biology, are produced by highly multivariate and nonlinear complex dynamical systems. In this paper, we focus on brain imaging data, including both calcium imaging and functional MRI data. Standard vector-autoregressive models are limited by their linearity assumptions, while nonlinear general-purpose, large-scale temporal models, such as LSTM networks, typically require large amounts of training data, not always readily available in biological applications; furthermore, such models have limited interpretability. We introduce here a novel approach for learning a nonlinear differential equation model aimed at capturing brain dynamics. Specifically, we propose a variable-projection optimization approach to estimate the parameters of the multivariate (coupled) van der Pol oscillator, and demonstrate that such a model can accurately represent nonlinear dynamics of the brain data. Furthermore, in order to improve the predictive accuracy when forecasting future brain-activity time series, we use this analytical model as an unlimited source of simulated data for pretraining LSTM; such model-specific data augmentation approach consistently improves LSTM performance on both calcium and fMRI imaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge