Oscar van der Heide
Quantitative mapping from conventional MRI using self-supervised physics-guided deep learning: applications to a large-scale, clinically heterogeneous dataset
Jan 08, 2026Abstract:Magnetic resonance imaging (MRI) is a cornerstone of clinical neuroimaging, yet conventional MRIs provide qualitative information heavily dependent on scanner hardware and acquisition settings. While quantitative MRI (qMRI) offers intrinsic tissue parameters, the requirement for specialized acquisition protocols and reconstruction algorithms restricts its availability and impedes large-scale biomarker research. This study presents a self-supervised physics-guided deep learning framework to infer quantitative T1, T2, and proton-density (PD) maps directly from widely available clinical conventional T1-weighted, T2-weighted, and FLAIR MRIs. The framework was trained and evaluated on a large-scale, clinically heterogeneous dataset comprising 4,121 scan sessions acquired at our institution over six years on four different 3 T MRI scanner systems, capturing real-world clinical variability. The framework integrates Bloch-based signal models directly into the training objective. Across more than 600 test sessions, the generated maps exhibited white matter and gray matter values consistent with literature ranges. Additionally, the generated maps showed invariance to scanner hardware and acquisition protocol groups, with inter-group coefficients of variation $\leq$ 1.1%. Subject-specific analyses demonstrated excellent voxel-wise reproducibility across scanner systems and sequence parameters, with Pearson $r$ and concordance correlation coefficients exceeding 0.82 for T1 and T2. Mean relative voxel-wise differences were low across all quantitative parameters, especially for T2 ($<$ 6%). These results indicate that the proposed framework can robustly transform diverse clinical conventional MRI data into quantitative maps, potentially paving the way for large-scale quantitative biomarker research.
Time-efficient, high-resolution 3T whole-brain relaxometry using Cartesian 3D MR-STAT with CSF suppression
Mar 22, 2024



Abstract:Purpose: Current 3D Magnetic Resonance Spin TomogrAphy in Time-domain (MR-STAT) protocols use transient-state, gradient-spoiled gradient-echo sequences that are prone to cerebrospinal fluid (CSF) pulsation artifacts when applied to the brain. This study aims at developing a 3D MR-STAT protocol for whole-brain relaxometry that overcomes the challenges posed by CSF-induced ghosting artifacts. Method: We optimized the flip-angle train within the Cartesian 3D MR-STAT framework to achieve two objectives: (1) minimization of the noise level in the reconstructed quantitative maps, and (2) reduction of the CSF-to-white-matter signal ratio to suppress CSF signal and the associated pulsation artifacts. The optimized new sequence was tested on a gel/water-phantom to evaluate the accuracy of the quantitative maps, and on healthy volunteers to explore the effectiveness of the CSF artifact suppression and robustness of the new protocol. Results: A new optimized sequence with both high parameter encoding capability and low CSF intensity was proposed and initially validated in the gel/water-phantom experiment. From in-vivo experiments with five volunteers, the proposed CSF-suppressed sequence shows no CSF ghosting artifacts and overall greatly improved image quality for all quantitative maps compared to the baseline sequence. Statistical analysis indicated low inter-subject and inter-scan variability for quantitative parameters in gray matter and white matter (1.6%-2.4% for T1 and 2.0%-4.6% for T2), demonstrating the robustness of the new sequence. Conclusion: We presented a new 3D MR-STAT sequence with CSF suppression that effectively eliminates CSF pulsation artifacts. The new sequence ensures consistently high-quality, 1mm^3 whole-brain relaxometry within a rapid 5.5-minute scan time.
Acceleration Strategies for MR-STAT: Achieving High-Resolution Reconstructions on a Desktop PC within 3 minutes
May 04, 2022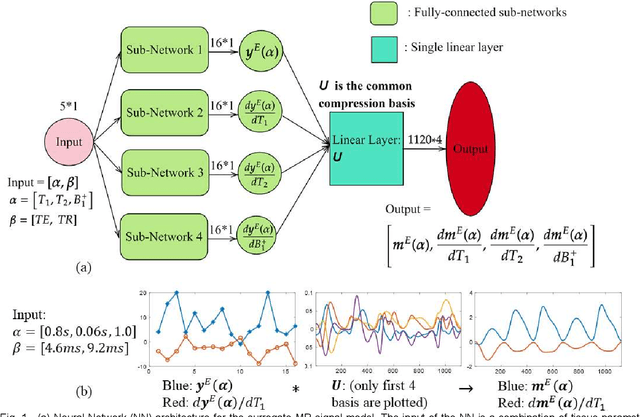
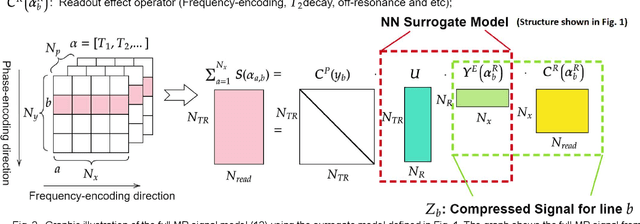
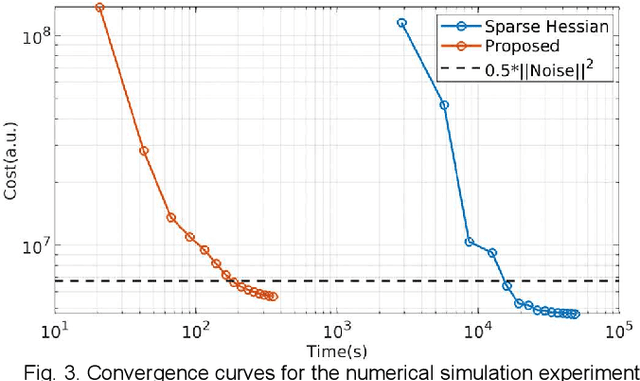
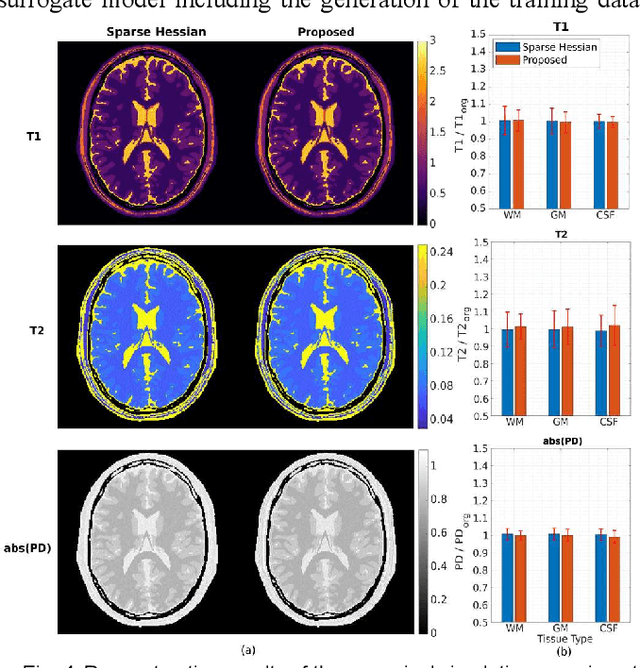
Abstract:MR-STAT is an emerging quantitative magnetic resonance imaging technique which aims at obtaining multi-parametric tissue parameter maps from single short scans. It describes the relationship between the spatial-domain tissue parameters and the time-domain measured signal by using a comprehensive, volumetric forward model. The MR-STAT reconstruction solves a large-scale nonlinear problem, thus is very computationally challenging. In previous work, MR-STAT reconstruction using Cartesian readout data was accelerated by approximating the Hessian matrix with sparse, banded blocks, and can be done on high performance CPU clusters with tens of minutes. In the current work, we propose an accelerated Cartesian MR-STAT algorithm incorporating two different strategies: firstly, a neural network is trained as a fast surrogate to learn the magnetization signal not only in the full time-domain but also in the compressed lowrank domain; secondly, based on the surrogate model, the Cartesian MR-STAT problem is re-formulated and split into smaller sub-problems by the alternating direction method of multipliers. The proposed method substantially reduces the computational requirements for runtime and memory. Simulated and in-vivo balanced MR-STAT experiments show similar reconstruction results using the proposed algorithm compared to the previous sparse Hessian method, and the reconstruction times are at least 40 times shorter. Incorporating sensitivity encoding and regularization terms is straightforward, and allows for better image quality with a negligible increase in reconstruction time. The proposed algorithm could reconstruct both balanced and gradient-spoiled in-vivo data within 3 minutes on a desktop PC, and could thereby facilitate the translation of MR-STAT in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge