Onur Boyar
LLM-Fusion: A Novel Multimodal Fusion Model for Accelerated Material Discovery
Mar 02, 2025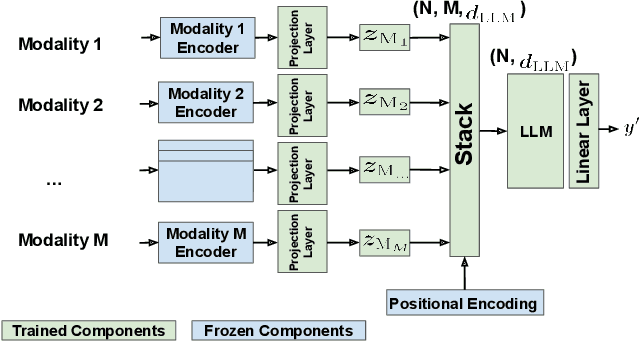
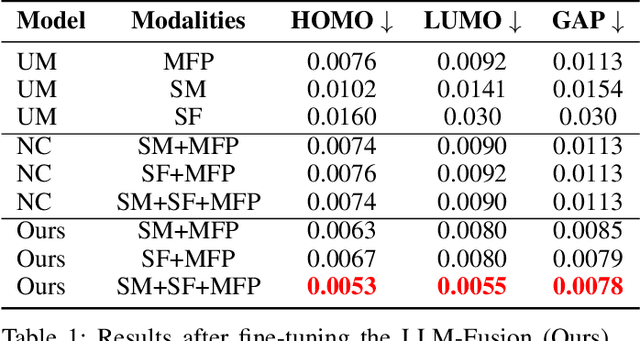
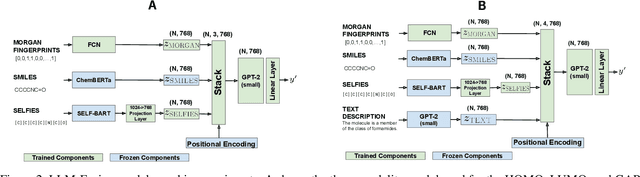
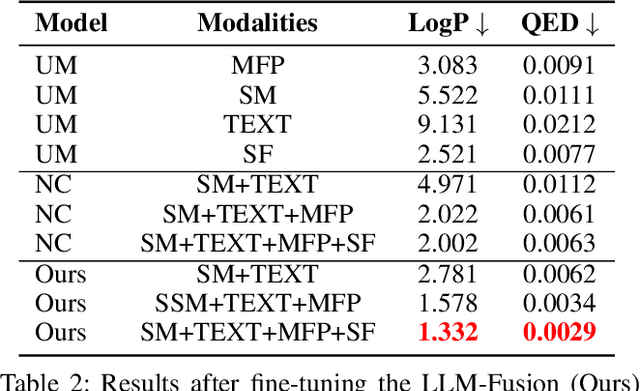
Abstract:Discovering materials with desirable properties in an efficient way remains a significant problem in materials science. Many studies have tackled this problem by using different sets of information available about the materials. Among them, multimodal approaches have been found to be promising because of their ability to combine different sources of information. However, fusion algorithms to date remain simple, lacking a mechanism to provide a rich representation of multiple modalities. This paper presents LLM-Fusion, a novel multimodal fusion model that leverages large language models (LLMs) to integrate diverse representations, such as SMILES, SELFIES, text descriptions, and molecular fingerprints, for accurate property prediction. Our approach introduces a flexible LLM-based architecture that supports multimodal input processing and enables material property prediction with higher accuracy than traditional methods. We validate our model on two datasets across five prediction tasks and demonstrate its effectiveness compared to unimodal and naive concatenation baselines.
Conditional Latent Space Molecular Scaffold Optimization for Accelerated Molecular Design
Nov 03, 2024Abstract:The rapid discovery of new chemical compounds is essential for advancing global health and developing treatments. While generative models show promise in creating novel molecules, challenges remain in ensuring the real-world applicability of these molecules and finding such molecules efficiently. To address this, we introduce Conditional Latent Space Molecular Scaffold Optimization (CLaSMO), which combines a Conditional Variational Autoencoder (CVAE) with Latent Space Bayesian Optimization (LSBO) to modify molecules strategically while maintaining similarity to the original input. Our LSBO setting improves the sample-efficiency of our optimization, and our modification approach helps us to obtain molecules with higher chances of real-world applicability. CLaSMO explores substructures of molecules in a sample-efficient manner by performing BO in the latent space of a CVAE conditioned on the atomic environment of the molecule to be optimized. Our experiments demonstrate that CLaSMO efficiently enhances target properties with minimal substructure modifications, achieving state-of-the-art results with a smaller model and dataset compared to existing methods. We also provide an open-source web application that enables chemical experts to apply CLaSMO in a Human-in-the-Loop setting.
Crystal-LSBO: Automated Design of De Novo Crystals with Latent Space Bayesian Optimization
May 28, 2024Abstract:Generative modeling of crystal structures is significantly challenged by the complexity of input data, which constrains the ability of these models to explore and discover novel crystals. This complexity often confines de novo design methodologies to merely small perturbations of known crystals and hampers the effective application of advanced optimization techniques. One such optimization technique, Latent Space Bayesian Optimization (LSBO) has demonstrated promising results in uncovering novel objects across various domains, especially when combined with Variational Autoencoders (VAEs). Recognizing LSBO's potential and the critical need for innovative crystal discovery, we introduce Crystal-LSBO, a de novo design framework for crystals specifically tailored to enhance explorability within LSBO frameworks. Crystal-LSBO employs multiple VAEs, each dedicated to a distinct aspect of crystal structure: lattice, coordinates, and chemical elements, orchestrated by an integrative model that synthesizes these components into a cohesive output. This setup not only streamlines the learning process but also produces explorable latent spaces thanks to the decreased complexity of the learning task for each model, enabling LSBO approaches to operate. Our study pioneers the use of LSBO for de novo crystal design, demonstrating its efficacy through optimization tasks focused mainly on formation energy values. Our results highlight the effectiveness of our methodology, offering a new perspective for de novo crystal discovery.
Latent Reconstruction-Aware Variational Autoencoder
Feb 05, 2023Abstract:Variational Autoencoders (VAEs) have become increasingly popular in recent years due to their ability to generate new objects such as images and texts from a given dataset. This ability has led to a wide range of applications. While standard tasks often require sampling from high-density regions in the latent space, there are also tasks that require sampling from low-density regions, such as Morphing and Latent Space Bayesian Optimization (LS-BO). These tasks are becoming increasingly important in fields such as de novo molecular design, where the ability to generate diverse and high-quality chemical compounds is essential. In this study, we investigate the issue of low-quality objects generated from low-density regions in VAEs. To address this problem, we propose a new VAE model, the Latent Reconstruction-Aware VAE (LRA-VAE). The LRA-VAE model takes into account what we refer to as the Latent Reconstruction Error (LRE) of the latent variables. We evaluate our proposal using Morphing and LS-BO experiments, and show that LRA-VAE can improve the quality of generated objects over the other approaches, making it a promising solution for various generation tasks that involve sampling from low-density regions.
Exploring The Limits Of Data Augmentation For Retinal Vessel Segmentation
May 30, 2021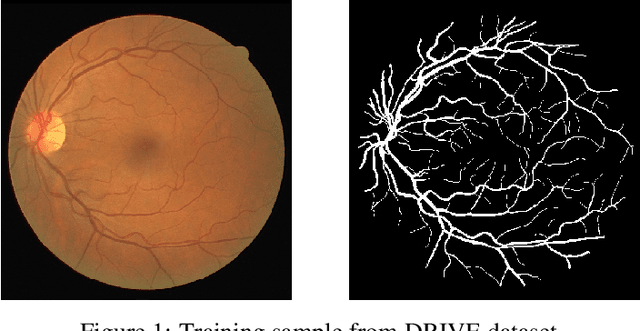
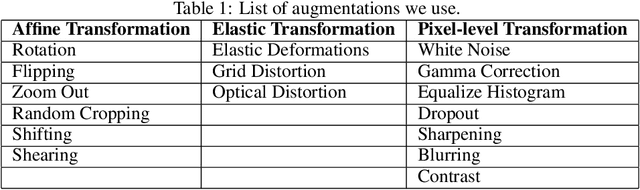
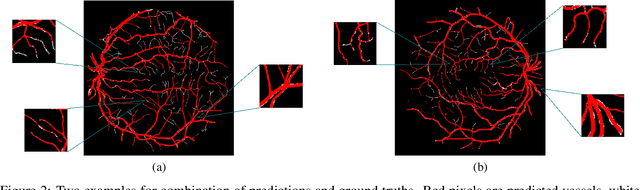
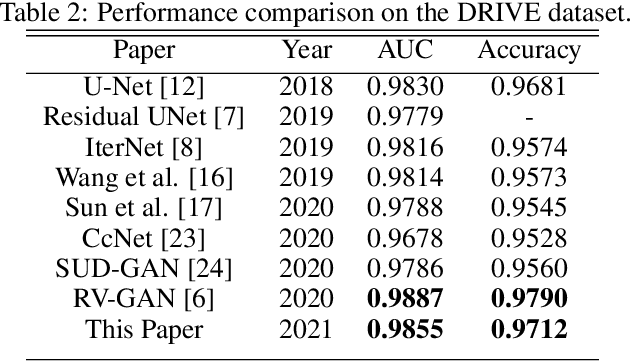
Abstract:Retinal Vessel Segmentation is important for the diagnosis of various diseases. The research on retinal vessel segmentation focuses mainly on the improvement of the segmentation model which is usually based on U-Net architecture. In our study, we use the U-Net architecture and we rely on heavy data augmentation in order to achieve better performance. The success of the data augmentation relies on successfully addressing the problem of input images. By analyzing input images and performing the augmentation accordingly we show that the performance of the U-Net model can be increased dramatically. Results are reported using the most widely used retina dataset, DRIVE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge