Omid Bazgir
Integration of Graph Neural Network and Neural-ODEs for Tumor Dynamic Prediction
Oct 02, 2023Abstract:In anti-cancer drug development, a major scientific challenge is disentangling the complex relationships between high-dimensional genomics data from patient tumor samples, the corresponding tumor's organ of origin, the drug targets associated with given treatments and the resulting treatment response. Furthermore, to realize the aspirations of precision medicine in identifying and adjusting treatments for patients depending on the therapeutic response, there is a need for building tumor dynamic models that can integrate both longitudinal tumor size as well as multimodal, high-content data. In this work, we take a step towards enhancing personalized tumor dynamic predictions by proposing a heterogeneous graph encoder that utilizes a bipartite Graph Convolutional Neural network (GCN) combined with Neural Ordinary Differential Equations (Neural-ODEs). We applied the methodology to a large collection of patient-derived xenograft (PDX) data, spanning a wide variety of treatments (as well as their combinations) on tumors that originated from a number of different organs. We first show that the methodology is able to discover a tumor dynamic model that significantly improves upon an empirical model which is in current use. Additionally, we show that the graph encoder is able to effectively utilize multimodal data to enhance tumor predictions. Our findings indicate that the methodology holds significant promise and offers potential applications in pre-clinical settings.
Investigation of REFINED CNN ensemble learning for anti-cancer drug sensitivity prediction
Sep 09, 2020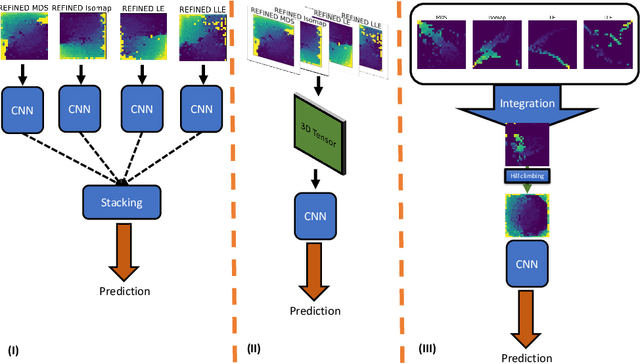
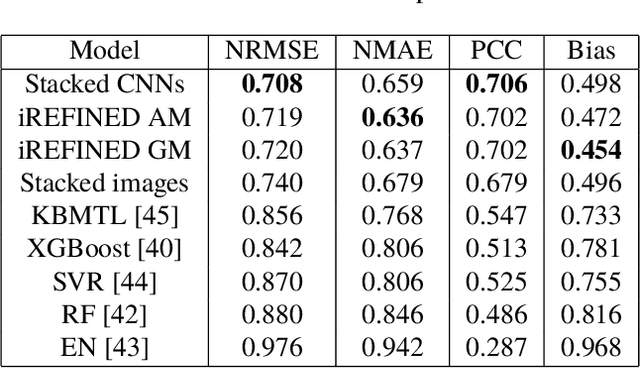

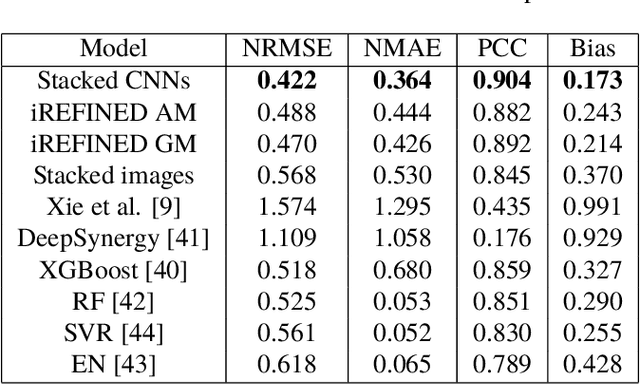
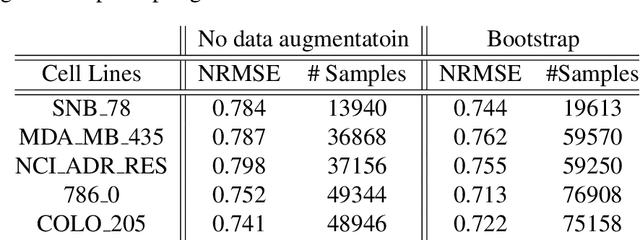
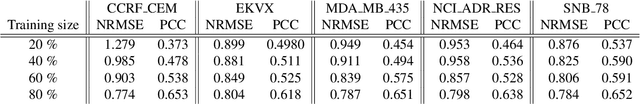
Abstract:Anti-cancer drug sensitivity prediction using deep learning models for individual cell line is a significant challenge in personalized medicine. REFINED (REpresentation of Features as Images with NEighborhood Dependencies) CNN (Convolutional Neural Network) based models have shown promising results in drug sensitivity prediction. The primary idea behind REFINED CNN is representing high dimensional vectors as compact images with spatial correlations that can benefit from convolutional neural network architectures. However, the mapping from a vector to a compact 2D image is not unique due to variations in considered distance measures and neighborhoods. In this article, we consider predictions based on ensembles built from such mappings that can improve upon the best single REFINED CNN model prediction. Results illustrated using NCI60 and NCIALMANAC databases shows that the ensemble approaches can provide significant performance improvement as compared to individual models. We further illustrate that a single mapping created from the amalgamation of the different mappings can provide performance similar to stacking ensemble but with significantly lower computational complexity.
Kidney segmentation using 3D U-Net localized with Expectation Maximization
Mar 20, 2020



Abstract:Kidney volume is greatly affected in several renal diseases. Precise and automatic segmentation of the kidney can help determine kidney size and evaluate renal function. Fully convolutional neural networks have been used to segment organs from large biomedical 3D images. While these networks demonstrate state-of-the-art segmentation performances, they do not immediately translate to small foreground objects, small sample sizes, and anisotropic resolution in MRI datasets. In this paper we propose a new framework to address some of the challenges for segmenting 3D MRI. These methods were implemented on preclinical MRI for segmenting kidneys in an animal model of lupus nephritis. Our implementation strategy is twofold: 1) to utilize additional MRI diffusion images to detect the general kidney area, and 2) to reduce the 3D U-Net kernels to handle small sample sizes. Using this approach, a Dice similarity coefficient of 0.88 was achieved with a limited dataset of n=196. This segmentation strategy with careful optimization can be applied to various renal injuries or other organ systems.
REFINED (REpresentation of Features as Images with NEighborhood Dependencies): A novel feature representation for Convolutional Neural Networks
Dec 11, 2019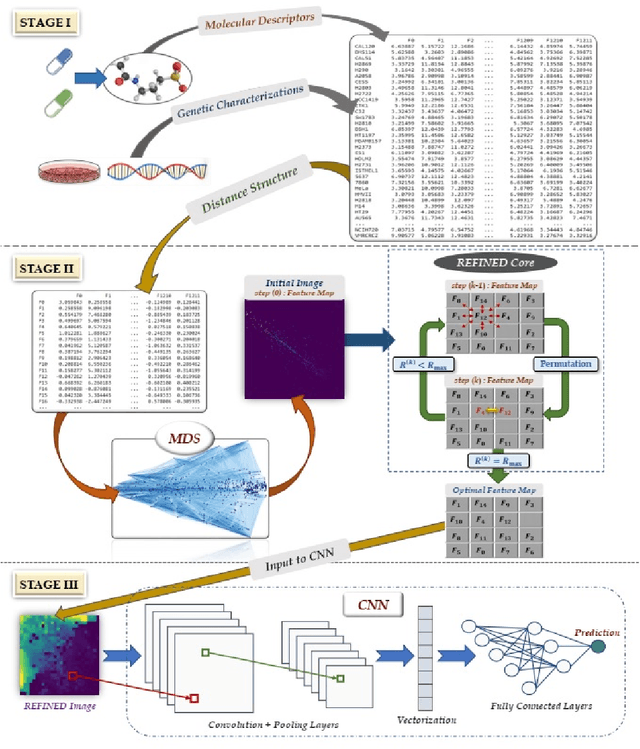
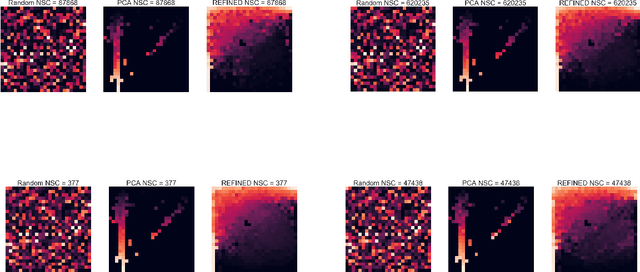
Abstract:Deep learning with Convolutional Neural Networks has shown great promise in various areas of image-based classification and enhancement but is often unsuitable for predictive modeling involving non-image based features or features without spatial correlations. We present a novel approach for representation of high dimensional feature vector in a compact image form, termed REFINED (REpresentation of Features as Images with NEighborhood Dependencies), that is conducible for convolutional neural network based deep learning. We consider the correlations between features to generate a compact representation of the features in the form of a two-dimensional image using minimization of pairwise distances similar to multi-dimensional scaling. We hypothesize that this approach enables embedded feature selection and integrated with Convolutional Neural Network based Deep Learning can produce more accurate predictions as compared to Artificial Neural Networks, Random Forests and Support Vector Regression. We illustrate the superior predictive performance of the proposed representation, as compared to existing approaches, using synthetic datasets, cell line efficacy prediction based on drug chemical descriptors for NCI60 dataset and drug sensitivity prediction based on transcriptomic data and chemical descriptors using GDSC dataset. Results illustrated on both synthetic and biological datasets shows the higher prediction accuracy of the proposed framework as compared to existing methodologies while maintaining desirable properties in terms of bias and feature extraction.
Emotion Recognition with Machine Learning Using EEG Signals
Mar 18, 2019



Abstract:In this research, an emotion recognition system is developed based on valence/arousal model using electroencephalography (EEG) signals. EEG signals are decomposed into the gamma, beta, alpha and theta frequency bands using discrete wavelet transform (DWT), and spectral features are extracted from each frequency band. Principle component analysis (PCA) is applied to the extracted features by preserving the same dimensionality, as a transform, to make the features mutually uncorrelated. Support vector machine (SVM), K-nearest neighbor (KNN) and artificial neural network (ANN) are used to classify emotional states. The cross-validated SVM with radial basis function (RBF) kernel using extracted features of 10 EEG channels, performs with 91.3% accuracy for arousal and 91.1% accuracy for valence, both in the beta frequency band. Our approach shows better performance compared to existing algorithms applied to the "DEAP" dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge