Omar Darwish
Leveraging Explainable AI for LLM Text Attribution: Differentiating Human-Written and Multiple LLMs-Generated Text
Jan 06, 2025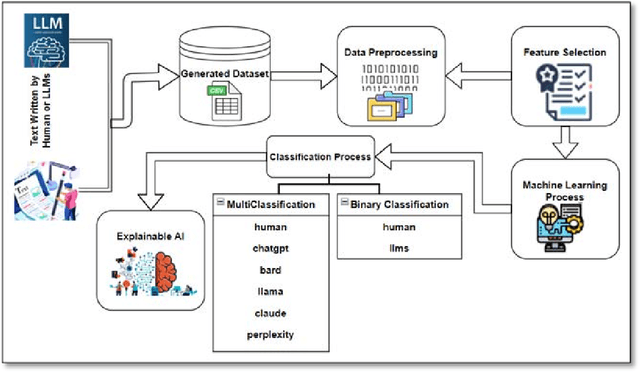
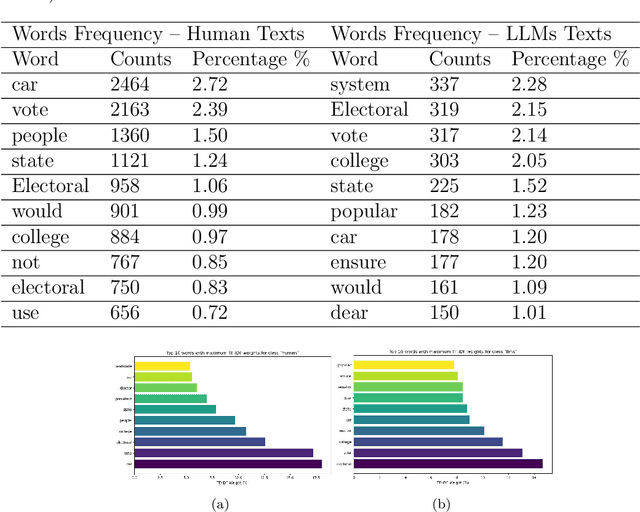
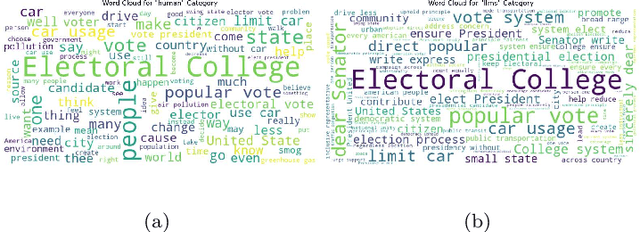
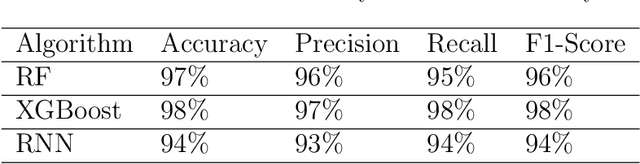
Abstract:The development of Generative AI Large Language Models (LLMs) raised the alarm regarding identifying content produced through generative AI or humans. In one case, issues arise when students heavily rely on such tools in a manner that can affect the development of their writing or coding skills. Other issues of plagiarism also apply. This study aims to support efforts to detect and identify textual content generated using LLM tools. We hypothesize that LLMs-generated text is detectable by machine learning (ML), and investigate ML models that can recognize and differentiate texts generated by multiple LLMs tools. We leverage several ML and Deep Learning (DL) algorithms such as Random Forest (RF), and Recurrent Neural Networks (RNN), and utilized Explainable Artificial Intelligence (XAI) to understand the important features in attribution. Our method is divided into 1) binary classification to differentiate between human-written and AI-text, and 2) multi classification, to differentiate between human-written text and the text generated by the five different LLM tools (ChatGPT, LLaMA, Google Bard, Claude, and Perplexity). Results show high accuracy in the multi and binary classification. Our model outperformed GPTZero with 98.5\% accuracy to 78.3\%. Notably, GPTZero was unable to recognize about 4.2\% of the observations, but our model was able to recognize the complete test dataset. XAI results showed that understanding feature importance across different classes enables detailed author/source profiles. Further, aiding in attribution and supporting plagiarism detection by highlighting unique stylistic and structural elements ensuring robust content originality verification.
Deep Bayesian Recurrent Neural Networks for Somatic Variant Calling in Cancer
Dec 06, 2019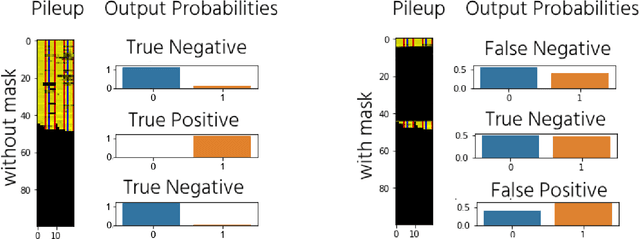
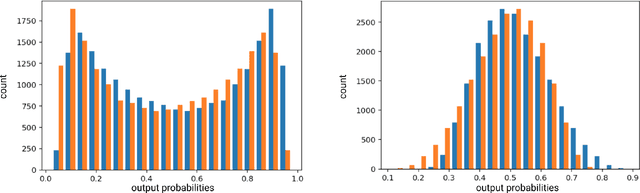
Abstract:The emerging field of precision oncology relies on the accurate pinpointing of alterations in the molecular profile of a tumor to provide personalized targeted treatments. Current methodologies in the field commonly include the application of next generation sequencing technologies to a tumor sample, followed by the identification of mutations in the DNA known as somatic variants. The differentiation of these variants from sequencing error poses a classic classification problem, which has traditionally been approached with Bayesian statistics, and more recently with supervised machine learning methods such as neural networks. Although these methods provide greater accuracy, classic neural networks lack the ability to indicate the confidence of a variant call. In this paper, we explore the performance of deep Bayesian neural networks on next generation sequencing data, and their ability to give probability estimates for somatic variant calls. In addition to demonstrating similar performance in comparison to standard neural networks, we show that the resultant output probabilities make these better suited to the disparate and highly-variable sequencing data-sets these models are likely to encounter in the real world. We aim to deliver algorithms to oncologists for which model certainty better reflects accuracy, for improved clinical application. By moving away from point estimates to reliable confidence intervals, we expect the resultant clinical and treatment decisions to be more robust and more informed by the underlying reality of the tumor molecular profile.
Safety and Robustness in Decision Making: Deep Bayesian Recurrent Neural Networks for Somatic Variant Calling in Cancer
Dec 04, 2019

Abstract:The genomic profile underlying an individual tumor can be highly informative in the creation of a personalized cancer treatment strategy for a given patient; a practice known as precision oncology. This involves next generation sequencing of a tumor sample and the subsequent identification of genomic aberrations, such as somatic mutations, to provide potential candidates of targeted therapy. The identification of these aberrations from sequencing noise and germline variant background poses a classic classification-style problem. This has been previously broached with many different supervised machine learning methods, including deep-learning neural networks. However, these neural networks have thus far not been tailored to give any indication of confidence in the mutation call, meaning an oncologist could be targeting a mutation with a low probability of being true. To address this, we present here a deep bayesian recurrent neural network for cancer variant calling, which shows no degradation in performance compared to standard neural networks. This approach enables greater flexibility through different priors to avoid overfitting to a single dataset. We will be incorporating this approach into software for oncologists to obtain safe, robust, and statistically confident somatic mutation calls for precision oncology treatment choices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge