Ohad Mosafi
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024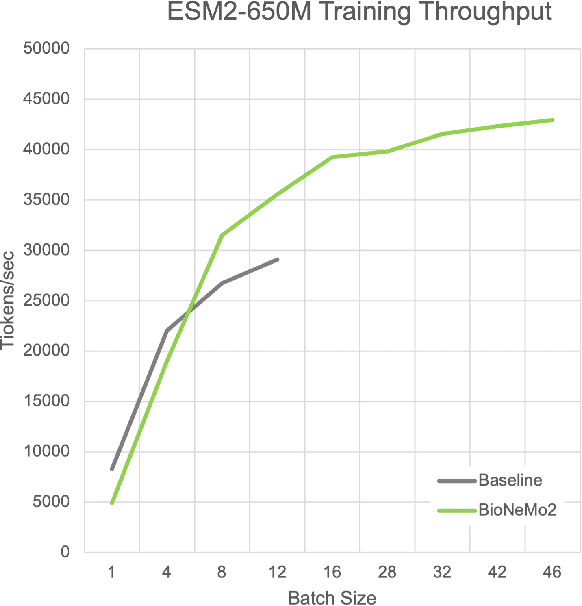
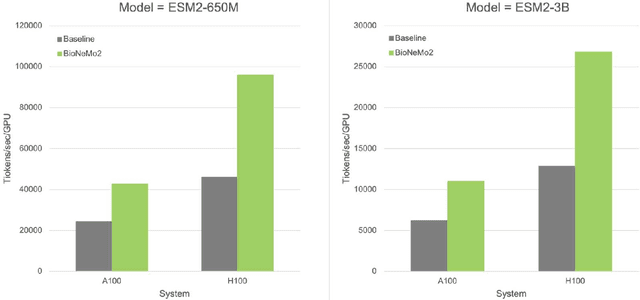
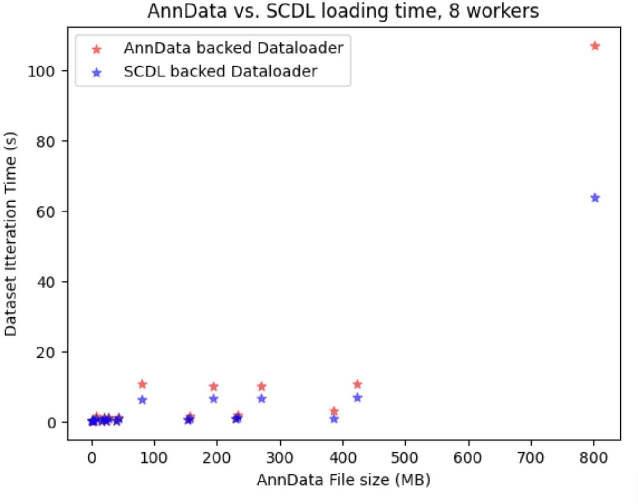
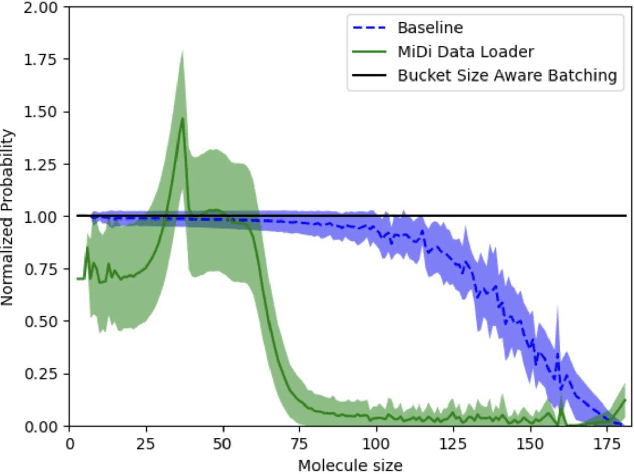
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
The UN Parallel Corpus Annotated for Translation Direction
May 20, 2018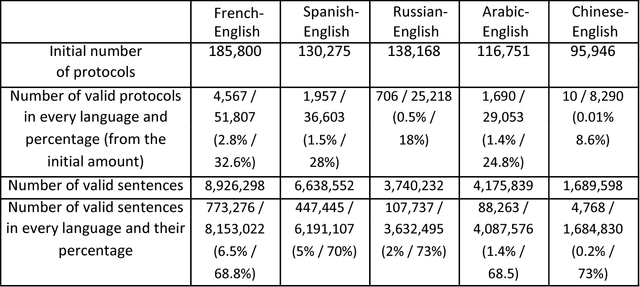
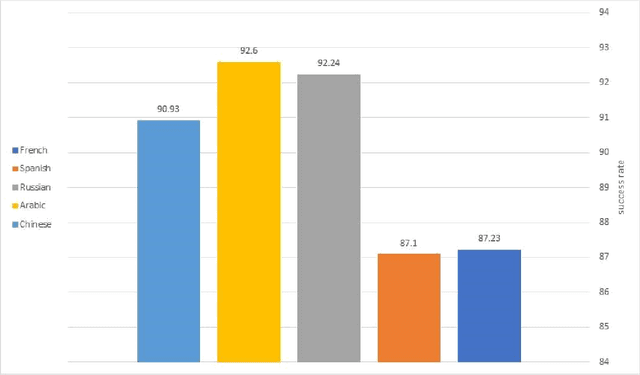
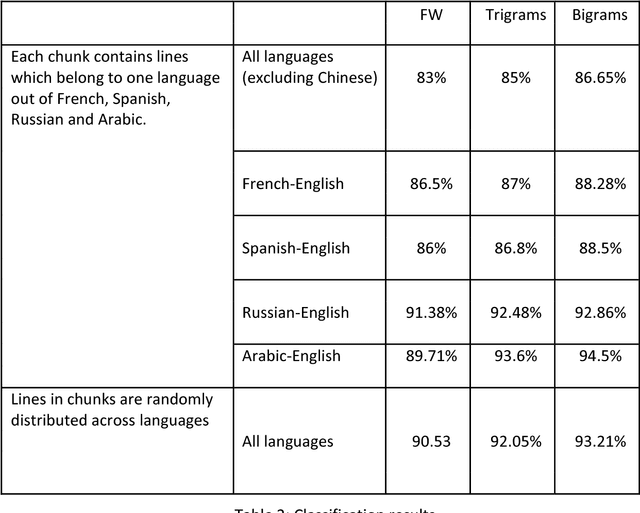

Abstract:This work distinguishes between translated and original text in the UN protocol corpus. By modeling the problem as classification problem, we can achieve up to 95% classification accuracy. We begin by deriving a parallel corpus for different language-pairs annotated for translation direction, and then classify the data by using various feature extraction methods. We compare the different methods as well as the ability to distinguish between translated and original texts in the different languages. The annotated corpus is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge