Nicolas Deutschmann
Conformal Autoregressive Generation: Beam Search with Coverage Guarantees
Sep 07, 2023Abstract:We introduce two new extensions to the beam search algorithm based on conformal predictions (CP) to produce sets of sequences with theoretical coverage guarantees. The first method is very simple and proposes dynamically-sized subsets of beam search results but, unlike typical CP procedures, has an upper bound on the achievable guarantee depending on a post-hoc calibration measure. Our second algorithm introduces the conformal set prediction procedure as part of the decoding process, producing a variable beam width which adapts to the current uncertainty. While more complex, this procedure can achieve coverage guarantees selected a priori. We provide marginal coverage bounds for each method, and evaluate them empirically on a selection of tasks drawing from natural language processing and chemistry.
Adaptive Conformal Regression with Jackknife+ Rescaled Scores
May 31, 2023



Abstract:Conformal regression provides prediction intervals with global coverage guarantees, but often fails to capture local error distributions, leading to non-homogeneous coverage. We address this with a new adaptive method based on rescaling conformal scores with an estimate of local score distribution, inspired by the Jackknife+ method, which enables the use of calibration data in conformal scores without breaking calibration-test exchangeability. Our approach ensures formal global coverage guarantees and is supported by new theoretical results on local coverage, including an a posteriori bound on any calibration score. The strength of our approach lies in achieving local coverage without sacrificing calibration set size, improving the applicability of conformal prediction intervals in various settings. As a result, our method provides prediction intervals that outperform previous methods, particularly in the low-data regime, making it especially relevant for real-world applications such as healthcare and biomedical domains where uncertainty needs to be quantified accurately despite low sample data.
Attention-based Interpretable Regression of Gene Expression in Histology
Aug 29, 2022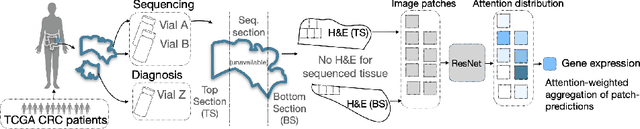
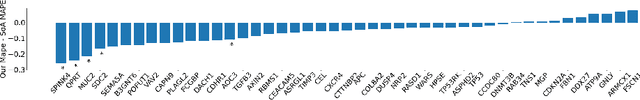


Abstract:Interpretability of deep learning is widely used to evaluate the reliability of medical imaging models and reduce the risks of inaccurate patient recommendations. For models exceeding human performance, e.g. predicting RNA structure from microscopy images, interpretable modelling can be further used to uncover highly non-trivial patterns which are otherwise imperceptible to the human eye. We show that interpretability can reveal connections between the microscopic appearance of cancer tissue and its gene expression profiling. While exhaustive profiling of all genes from the histology images is still challenging, we estimate the expression values of a well-known subset of genes that is indicative of cancer molecular subtype, survival, and treatment response in colorectal cancer. Our approach successfully identifies meaningful information from the image slides, highlighting hotspots of high gene expression. Our method can help characterise how gene expression shapes tissue morphology and this may be beneficial for patient stratification in the pathology unit. The code is available on GitHub.
Is Attention Interpretation? A Quantitative Assessment On Sets
Jul 26, 2022



Abstract:The debate around the interpretability of attention mechanisms is centered on whether attention scores can be used as a proxy for the relative amounts of signal carried by sub-components of data. We propose to study the interpretability of attention in the context of set machine learning, where each data point is composed of an unordered collection of instances with a global label. For classical multiple-instance-learning problems and simple extensions, there is a well-defined "importance" ground truth that can be leveraged to cast interpretation as a binary classification problem, which we can quantitatively evaluate. By building synthetic datasets over several data modalities, we perform a systematic assessment of attention-based interpretations. We find that attention distributions are indeed often reflective of the relative importance of individual instances, but that silent failures happen where a model will have high classification performance but attention patterns that do not align with expectations. Based on these observations, we propose to use ensembling to minimize the risk of misleading attention-based explanations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge