Nathan Cahill
Robust Spatial Filtering with Graph Convolutional Neural Networks
Jul 14, 2017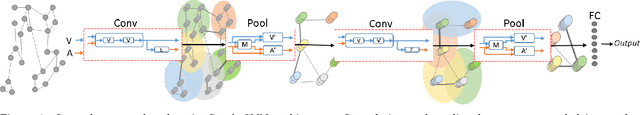
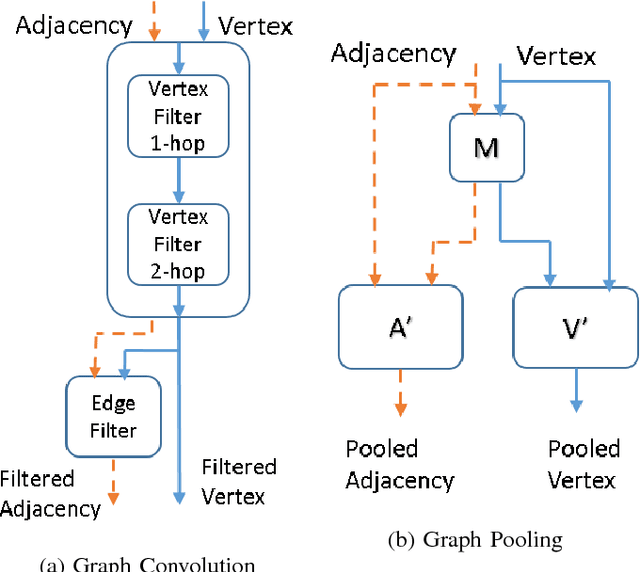
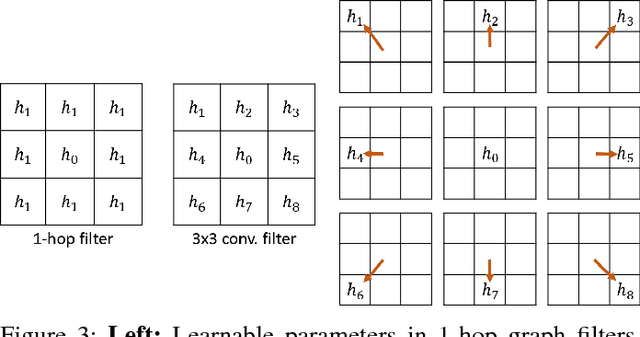
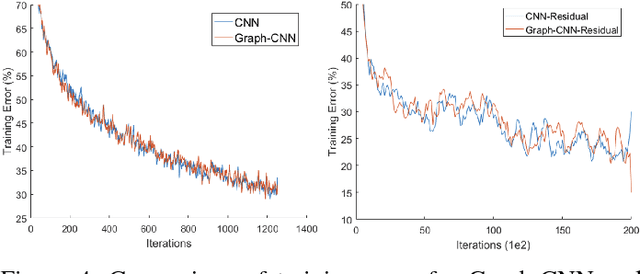
Abstract:Convolutional Neural Networks (CNNs) have recently led to incredible breakthroughs on a variety of pattern recognition problems. Banks of finite impulse response filters are learned on a hierarchy of layers, each contributing more abstract information than the previous layer. The simplicity and elegance of the convolutional filtering process makes them perfect for structured problems such as image, video, or voice, where vertices are homogeneous in the sense of number, location, and strength of neighbors. The vast majority of classification problems, for example in the pharmaceutical, homeland security, and financial domains are unstructured. As these problems are formulated into unstructured graphs, the heterogeneity of these problems, such as number of vertices, number of connections per vertex, and edge strength, cannot be tackled with standard convolutional techniques. We propose a novel neural learning framework that is capable of handling both homogeneous and heterogeneous data, while retaining the benefits of traditional CNN successes. Recently, researchers have proposed variations of CNNs that can handle graph data. In an effort to create learnable filter banks of graphs, these methods either induce constraints on the data or require preprocessing. As opposed to spectral methods, our framework, which we term Graph-CNNs, defines filters as polynomials of functions of the graph adjacency matrix. Graph-CNNs can handle both heterogeneous and homogeneous graph data, including graphs having entirely different vertex or edge sets. We perform experiments to validate the applicability of Graph-CNNs to a variety of structured and unstructured classification problems and demonstrate state-of-the-art results on document and molecule classification problems.
Integrating Atlas and Graph Cut Methods for LV Segmentation from Cardiac Cine MRI
Nov 03, 2016



Abstract:Magnetic Resonance Imaging (MRI) has evolved as a clinical standard-of-care imaging modality for cardiac morphology, function assessment, and guidance of cardiac interventions. All these applications rely on accurate extraction of the myocardial tissue and blood pool from the imaging data. Here we propose a framework for left ventricle (LV) segmentation from cardiac cine-MRI. First, we segment the LV blood pool using iterative graph cuts, and subsequently use this information to segment the myocardium. We formulate the segmentation procedure as an energy minimization problem in a graph subject to the shape prior obtained by label propagation from an average atlas using affine registration. The proposed framework has been validated on 30 patient cardiac cine-MRI datasets available through the STACOM LV segmentation challenge and yielded fast, robust, and accurate segmentation results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge