Naimul Mefraz Khan
DLIME: A Deterministic Local Interpretable Model-Agnostic Explanations Approach for Computer-Aided Diagnosis Systems
Jun 24, 2019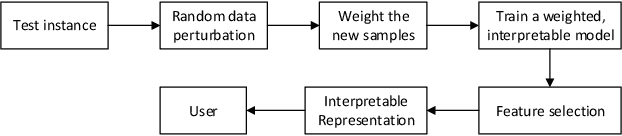
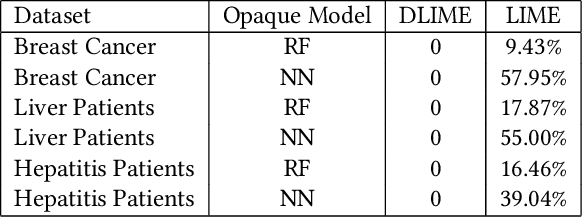
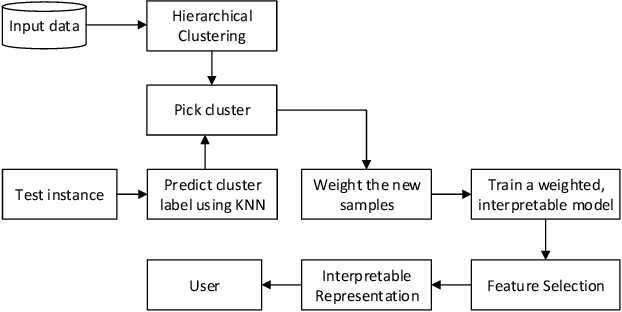
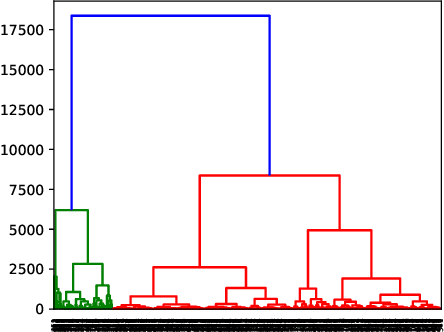
Abstract:Local Interpretable Model-Agnostic Explanations (LIME) is a popular technique used to increase the interpretability and explainability of black box Machine Learning (ML) algorithms. LIME typically generates an explanation for a single prediction by any ML model by learning a simpler interpretable model (e.g. linear classifier) around the prediction through generating simulated data around the instance by random perturbation, and obtaining feature importance through applying some form of feature selection. While LIME and similar local algorithms have gained popularity due to their simplicity, the random perturbation and feature selection methods result in "instability" in the generated explanations, where for the same prediction, different explanations can be generated. This is a critical issue that can prevent deployment of LIME in a Computer-Aided Diagnosis (CAD) system, where stability is of utmost importance to earn the trust of medical professionals. In this paper, we propose a deterministic version of LIME. Instead of random perturbation, we utilize agglomerative Hierarchical Clustering (HC) to group the training data together and K-Nearest Neighbour (KNN) to select the relevant cluster of the new instance that is being explained. After finding the relevant cluster, a linear model is trained over the selected cluster to generate the explanations. Experimental results on three different medical datasets show the superiority for Deterministic Local Interpretable Model-Agnostic Explanations (DLIME), where we quantitatively determine the stability of DLIME compared to LIME utilizing the Jaccard similarity among multiple generated explanations.
Transfer Learning with intelligent training data selection for prediction of Alzheimer's Disease
Jun 04, 2019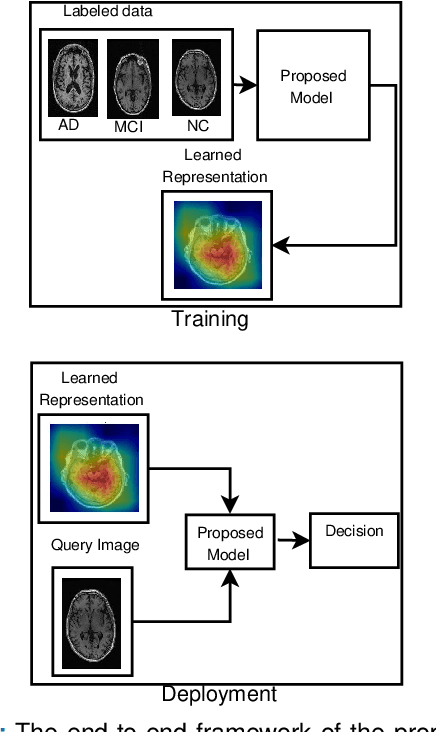
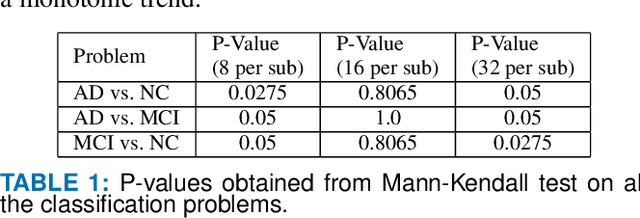
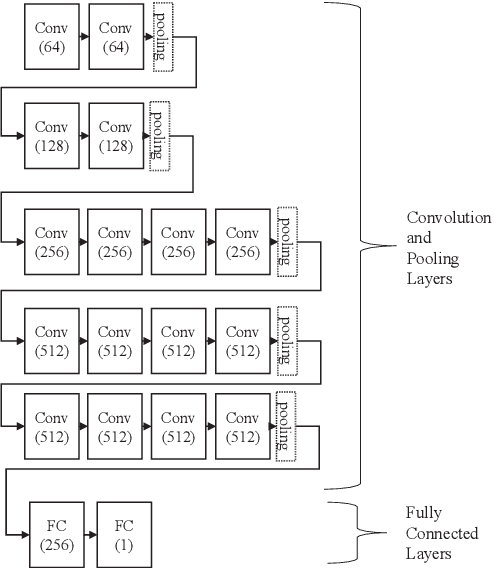
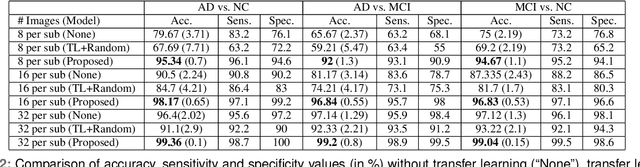
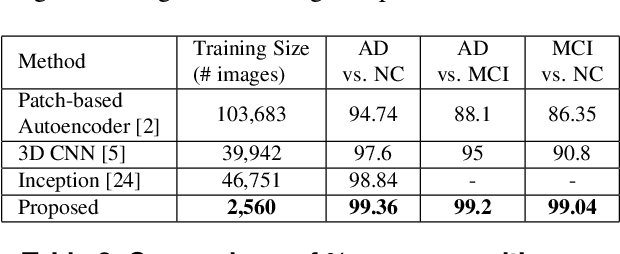
Abstract:Detection of Alzheimer's Disease (AD) from neuroimaging data such as MRI through machine learning has been a subject of intense research in recent years. Recent success of deep learning in computer vision has progressed such research further. However, common limitations with such algorithms are reliance on a large number of training images, and requirement of careful optimization of the architecture of deep networks. In this paper, we attempt solving these issues with transfer learning, where the state-of-the-art VGG architecture is initialized with pre-trained weights from large benchmark datasets consisting of natural images. The network is then fine-tuned with layer-wise tuning, where only a pre-defined group of layers are trained on MRI images. To shrink the training data size, we employ image entropy to select the most informative slices. Through experimentation on the ADNI dataset, we show that with training size of 10 to 20 times smaller than the other contemporary methods, we reach state-of-the-art performance in AD vs. NC, AD vs. MCI, and MCI vs. NC classification problems, with a 4% and a 7% increase in accuracy over the state-of-the-art for AD vs. MCI and MCI vs. NC, respectively. We also provide detailed analysis of the effect of the intelligent training data selection method, changing the training size, and changing the number of layers to be fine-tuned. Finally, we provide Class Activation Maps (CAM) that demonstrate how the proposed model focuses on discriminative image regions that are neuropathologically relevant, and can help the healthcare practitioner in interpreting the model's decision making process.
Machine Learning on Biomedical Images: Interactive Learning, Transfer Learning, Class Imbalance, and Beyond
Feb 13, 2019

Abstract:In this paper, we highlight three issues that limit performance of machine learning on biomedical images, and tackle them through 3 case studies: 1) Interactive Machine Learning (IML): we show how IML can drastically improve exploration time and quality of direct volume rendering. 2) transfer learning: we show how transfer learning along with intelligent pre-processing can result in better Alzheimer's diagnosis using a much smaller training set 3) data imbalance: we show how our novel focal Tversky loss function can provide better segmentation results taking into account the imbalanced nature of segmentation datasets. The case studies are accompanied by in-depth analytical discussion of results with possible future directions.
Deep Clustering with a Dynamic Autoencoder
Jan 23, 2019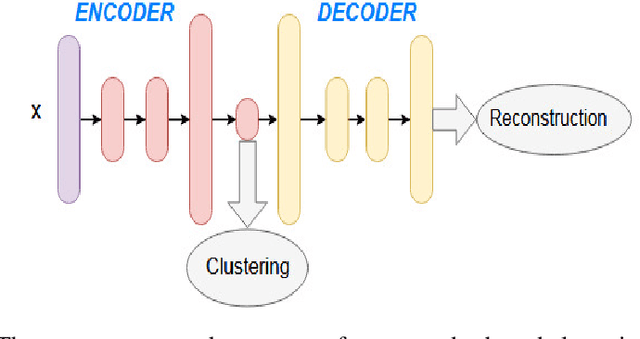


Abstract:In unsupervised learning, there is no obvious straightforward loss function which can capture the major factors of variations and similarities. Since natural systems have smooth dynamics, an opportunity is lost if an unsupervised loss function remains static during the training process. The absence of concrete supervision suggests that smooth complex dynamics should be integrated as a substitute to the classical static loss functions to better make use of the gradual and uncertain knowledge acquired through self-supervision. In this paper, we propose Dynamic Autoencoder (DynAE), a new model for deep clustering that allows to solve a clustering-reconstruction trade-off by gradually and smoothly eliminating the reconstruction objective in favor of a construction one while preserving the space topology. Experimental evaluations on benchmark datasets show that our approach achieves state-of-the-art results compared to all the other autoencoder-based clustering methods.
A Novel Focal Tversky loss function with improved Attention U-Net for lesion segmentation
Oct 18, 2018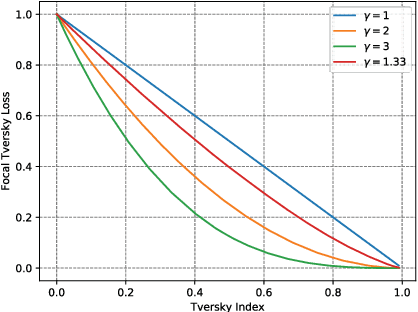
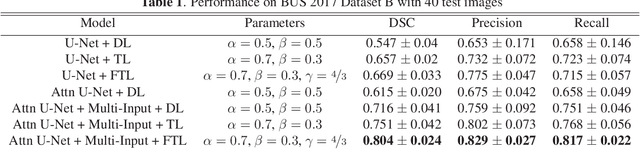
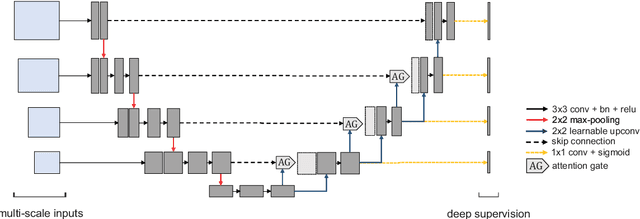
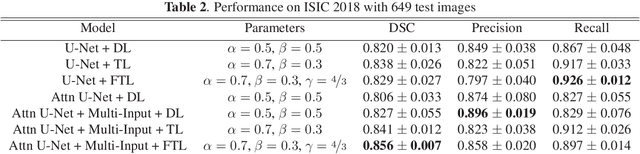
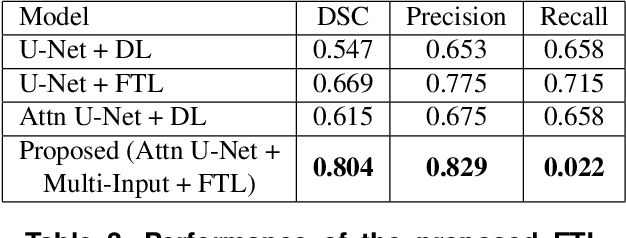
Abstract:We propose a generalized focal loss function based on the Tversky index to address the issue of data imbalance in medical image segmentation. Compared to the commonly used Dice loss, our loss function achieves a better trade off between precision and recall when training on small structures such as lesions. To evaluate our loss function, we improve the attention U-Net model by incorporating an image pyramid to preserve contextual features. We experiment on the BUS 2017 dataset and ISIC 2018 dataset where lesions occupy 4.84% and 21.4% of the images area and improve segmentation accuracy when compared to the standard U-Net by 25.7% and 3.6%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge