Naimat Ullah
CAMP: A Context-Aware Cricket Players Performance Metric
Jul 14, 2023Abstract:Cricket is the second most popular sport after soccer in terms of viewership. However, the assessment of individual player performance, a fundamental task in team sports, is currently primarily based on aggregate performance statistics, including average runs and wickets taken. We propose Context-Aware Metric of player Performance, CAMP, to quantify individual players' contributions toward a cricket match outcome. CAMP employs data mining methods and enables effective data-driven decision-making for selection and drafting, coaching and training, team line-ups, and strategy development. CAMP incorporates the exact context of performance, such as opponents' strengths and specific circumstances of games, such as pressure situations. We empirically evaluate CAMP on data of limited-over cricket matches between 2001 and 2019. In every match, a committee of experts declares one player as the best player, called Man of the M}atch (MoM). The top two rated players by CAMP match with MoM in 83\% of the 961 games. Thus, the CAMP rating of the best player closely matches that of the domain experts. By this measure, CAMP significantly outperforms the current best-known players' contribution measure based on the Duckworth-Lewis-Stern (DLS) method.
A k-mer Based Approach for SARS-CoV-2 Variant Identification
Aug 25, 2021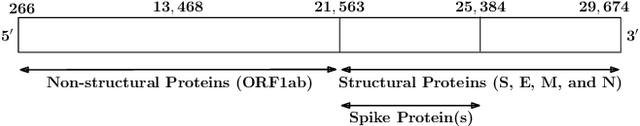
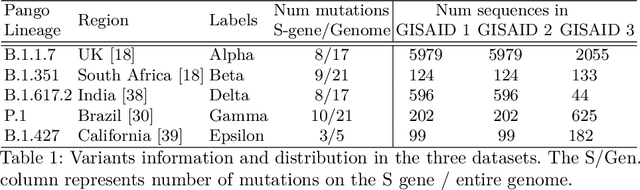
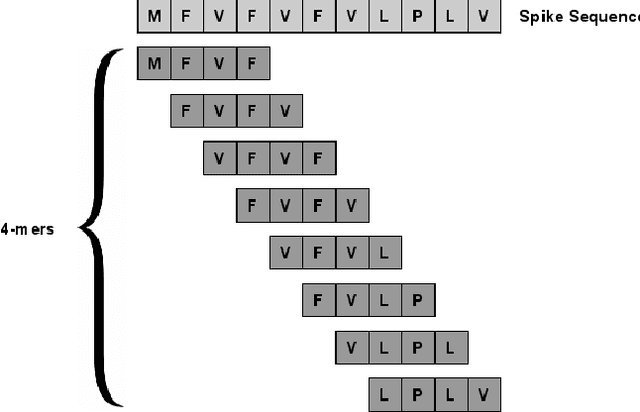
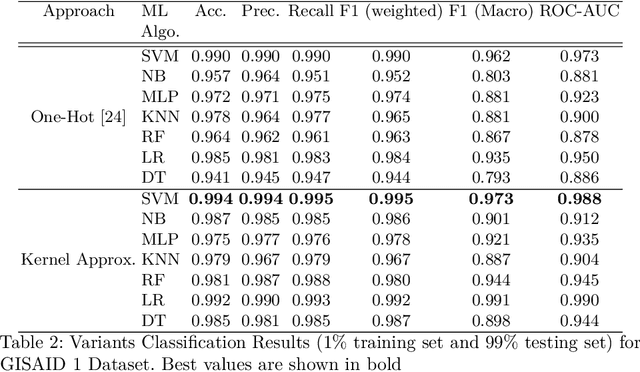
Abstract:With the rapid spread of the novel coronavirus (COVID-19) across the globe and its continuous mutation, it is of pivotal importance to design a system to identify different known (and unknown) variants of SARS-CoV-2. Identifying particular variants helps to understand and model their spread patterns, design effective mitigation strategies, and prevent future outbreaks. It also plays a crucial role in studying the efficacy of known vaccines against each variant and modeling the likelihood of breakthrough infections. It is well known that the spike protein contains most of the information/variation pertaining to coronavirus variants. In this paper, we use spike sequences to classify different variants of the coronavirus in humans. We show that preserving the order of the amino acids helps the underlying classifiers to achieve better performance. We also show that we can train our model to outperform the baseline algorithms using only a small number of training samples ($1\%$ of the data). Finally, we show the importance of the different amino acids which play a key role in identifying variants and how they coincide with those reported by the USA's Centers for Disease Control and Prevention (CDC).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge