Myunghwan Kim
Discovering Hypernymy in Text-Rich Heterogeneous Information Network by Exploiting Context Granularity
Sep 04, 2019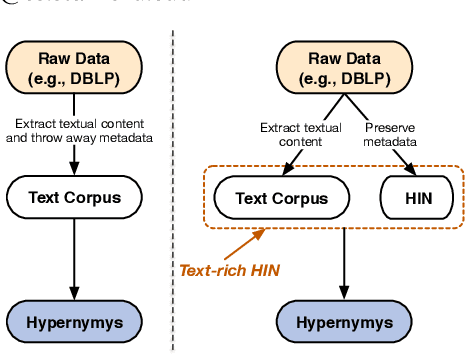
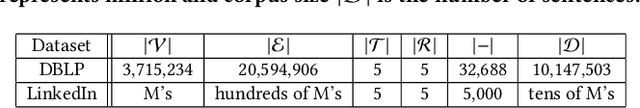
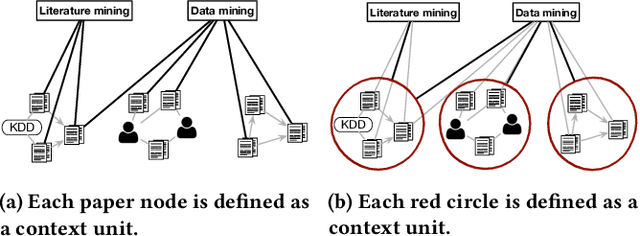
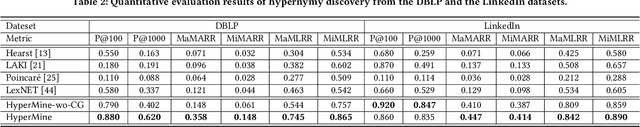
Abstract:Text-rich heterogeneous information networks (text-rich HINs) are ubiquitous in real-world applications. Hypernymy, also known as is-a relation or subclass-of relation, lays in the core of many knowledge graphs and benefits many downstream applications. Existing methods of hypernymy discovery either leverage textual patterns to extract explicitly mentioned hypernym-hyponym pairs, or learn a distributional representation for each term of interest based its context. These approaches rely on statistical signals from the textual corpus, and their effectiveness would therefore be hindered when the signals from the corpus are not sufficient for all terms of interest. In this work, we propose to discover hypernymy in text-rich HINs, which can introduce additional high-quality signals. We develop a new framework, named HyperMine, that exploits multi-granular contexts and combines signals from both text and network without human labeled data. HyperMine extends the definition of context to the scenario of text-rich HIN. For example, we can define typed nodes and communities as contexts. These contexts encode signals of different granularities and we feed them into a hypernymy inference model. HyperMine learns this model using weak supervision acquired based on high-precision textual patterns. Extensive experiments on two large real-world datasets demonstrate the effectiveness of HyperMine and the utility of modeling context granularity. We further show a case study that a high-quality taxonomy can be generated solely based on the hypernymy discovered by HyperMine.
Nonparametric Multi-group Membership Model for Dynamic Networks
Nov 08, 2013



Abstract:Relational data-like graphs, networks, and matrices-is often dynamic, where the relational structure evolves over time. A fundamental problem in the analysis of time-varying network data is to extract a summary of the common structure and the dynamics of the underlying relations between the entities. Here we build on the intuition that changes in the network structure are driven by the dynamics at the level of groups of nodes. We propose a nonparametric multi-group membership model for dynamic networks. Our model contains three main components: We model the birth and death of individual groups with respect to the dynamics of the network structure via a distance dependent Indian Buffet Process. We capture the evolution of individual node group memberships via a Factorial Hidden Markov model. And, we explain the dynamics of the network structure by explicitly modeling the connectivity structure of groups. We demonstrate our model's capability of identifying the dynamics of latent groups in a number of different types of network data. Experimental results show that our model provides improved predictive performance over existing dynamic network models on future network forecasting and missing link prediction.
Latent Multi-group Membership Graph Model
May 21, 2012



Abstract:We develop the Latent Multi-group Membership Graph (LMMG) model, a model of networks with rich node feature structure. In the LMMG model, each node belongs to multiple groups and each latent group models the occurrence of links as well as the node feature structure. The LMMG can be used to summarize the network structure, to predict links between the nodes, and to predict missing features of a node. We derive efficient inference and learning algorithms and evaluate the predictive performance of the LMMG on several social and document network datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge