Muhammad Moazam Fraz
'Aariz: A Benchmark Dataset for Automatic Cephalometric Landmark Detection and CVM Stage Classification
Feb 15, 2023



Abstract:The accurate identification and precise localization of cephalometric landmarks enable the classification and quantification of anatomical abnormalities. The traditional way of marking cephalometric landmarks on lateral cephalograms is a monotonous and time-consuming job. Endeavours to develop automated landmark detection systems have persistently been made, however, they are inadequate for orthodontic applications due to unavailability of a reliable dataset. We proposed a new state-of-the-art dataset to facilitate the development of robust AI solutions for quantitative morphometric analysis. The dataset includes 1000 lateral cephalometric radiographs (LCRs) obtained from 7 different radiographic imaging devices with varying resolutions, making it the most diverse and comprehensive cephalometric dataset to date. The clinical experts of our team meticulously annotated each radiograph with 29 cephalometric landmarks, including the most significant soft tissue landmarks ever marked in any publicly available dataset. Additionally, our experts also labelled the cervical vertebral maturation (CVM) stage of the patient in a radiograph, making this dataset the first standard resource for CVM classification. We believe that this dataset will be instrumental in the development of reliable automated landmark detection frameworks for use in orthodontics and beyond.
CEPHA29: Automatic Cephalometric Landmark Detection Challenge 2023
Dec 09, 2022



Abstract:Quantitative cephalometric analysis is the most widely used clinical and research tool in modern orthodontics. Accurate localization of cephalometric landmarks enables the quantification and classification of anatomical abnormalities, however, the traditional manual way of marking these landmarks is a very tedious job. Endeavours have constantly been made to develop automated cephalometric landmark detection systems but they are inadequate for orthodontic applications. The fundamental reason for this is that the amount of publicly available datasets as well as the images provided for training in these datasets are insufficient for an AI model to perform well. To facilitate the development of robust AI solutions for morphometric analysis, we organise the CEPHA29 Automatic Cephalometric Landmark Detection Challenge in conjunction with IEEE International Symposium on Biomedical Imaging (ISBI 2023). In this context, we provide the largest known publicly available dataset, consisting of 1000 cephalometric X-ray images. We hope that our challenge will not only derive forward research and innovation in automatic cephalometric landmark identification but will also signal the beginning of a new era in the discipline.
HistoSeg : Quick attention with multi-loss function for multi-structure segmentation in digital histology images
Sep 01, 2022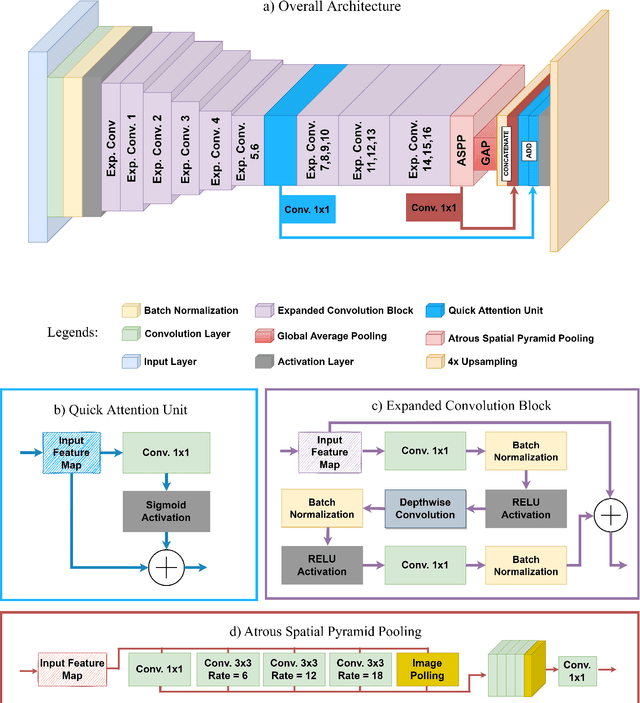
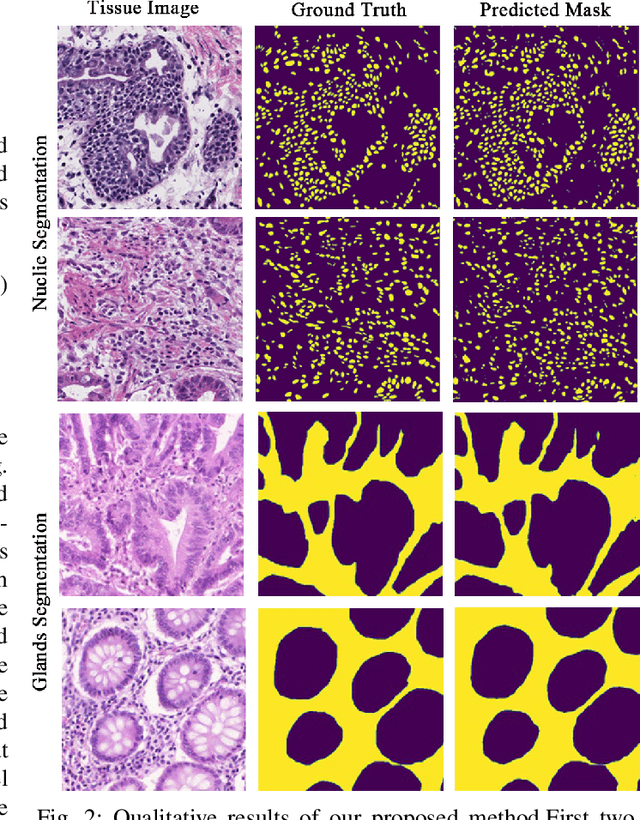
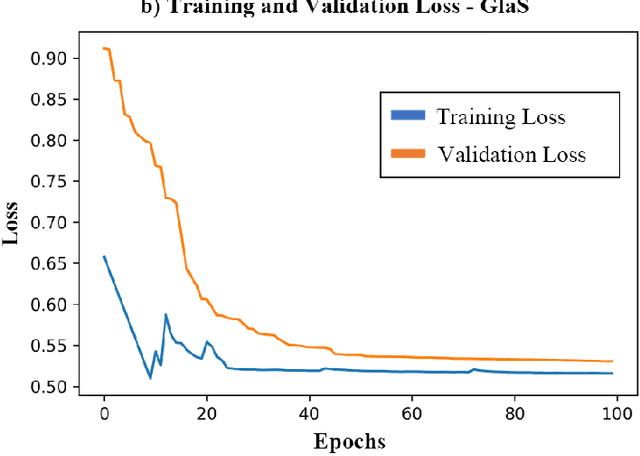

Abstract:Medical image segmentation assists in computer-aided diagnosis, surgeries, and treatment. Digitize tissue slide images are used to analyze and segment glands, nuclei, and other biomarkers which are further used in computer-aided medical applications. To this end, many researchers developed different neural networks to perform segmentation on histological images, mostly these networks are based on encoder-decoder architecture and also utilize complex attention modules or transformers. However, these networks are less accurate to capture relevant local and global features with accurate boundary detection at multiple scales, therefore, we proposed an Encoder-Decoder Network, Quick Attention Module and a Multi Loss Function (combination of Binary Cross Entropy (BCE) Loss, Focal Loss & Dice Loss). We evaluate the generalization capability of our proposed network on two publicly available datasets for medical image segmentation MoNuSeg and GlaS and outperform the state-of-the-art networks with 1.99% improvement on the MoNuSeg dataset and 7.15% improvement on the GlaS dataset. Implementation Code is available at this link: https://bit.ly/HistoSeg
Nuclei & Glands Instance Segmentation in Histology Images: A Narrative Review
Aug 26, 2022

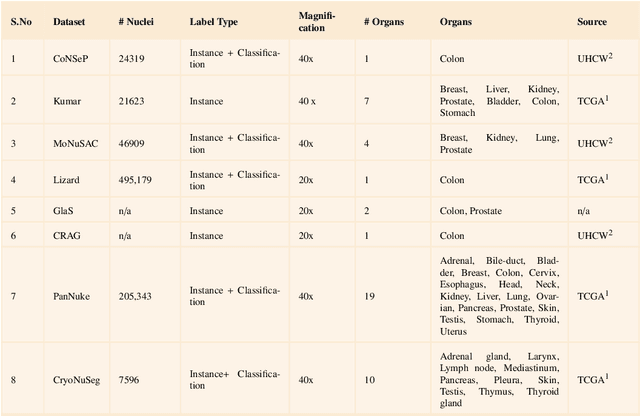
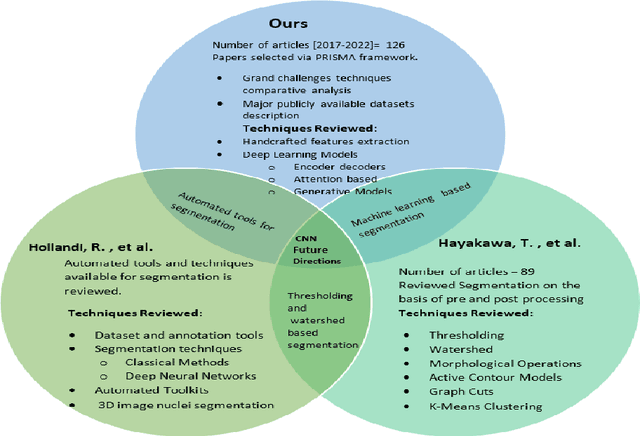
Abstract:Instance segmentation of nuclei and glands in the histology images is an important step in computational pathology workflow for cancer diagnosis, treatment planning and survival analysis. With the advent of modern hardware, the recent availability of large-scale quality public datasets and the community organized grand challenges have seen a surge in automated methods focusing on domain specific challenges, which is pivotal for technology advancements and clinical translation. In this survey, 126 papers illustrating the AI based methods for nuclei and glands instance segmentation published in the last five years (2017-2022) are deeply analyzed, the limitations of current approaches and the open challenges are discussed. Moreover, the potential future research direction is presented and the contribution of state-of-the-art methods is summarized. Further, a generalized summary of publicly available datasets and a detailed insights on the grand challenges illustrating the top performing methods specific to each challenge is also provided. Besides, we intended to give the reader current state of existing research and pointers to the future directions in developing methods that can be used in clinical practice enabling improved diagnosis, grading, prognosis, and treatment planning of cancer. To the best of our knowledge, no previous work has reviewed the instance segmentation in histology images focusing towards this direction.
Vision Transformers in Medical Computer Vision -- A Contemplative Retrospection
Mar 29, 2022
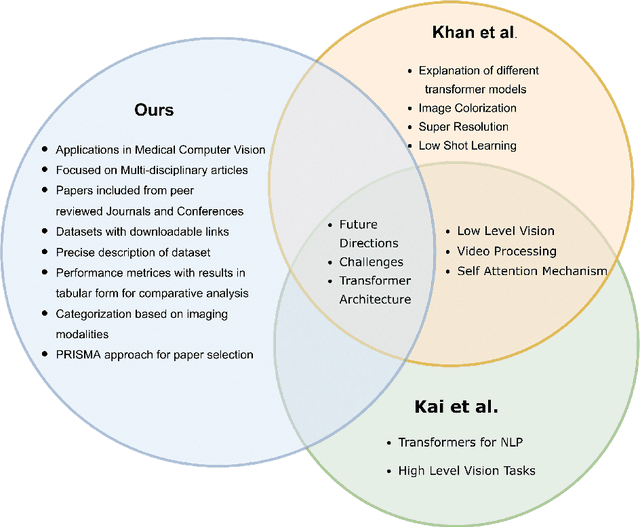
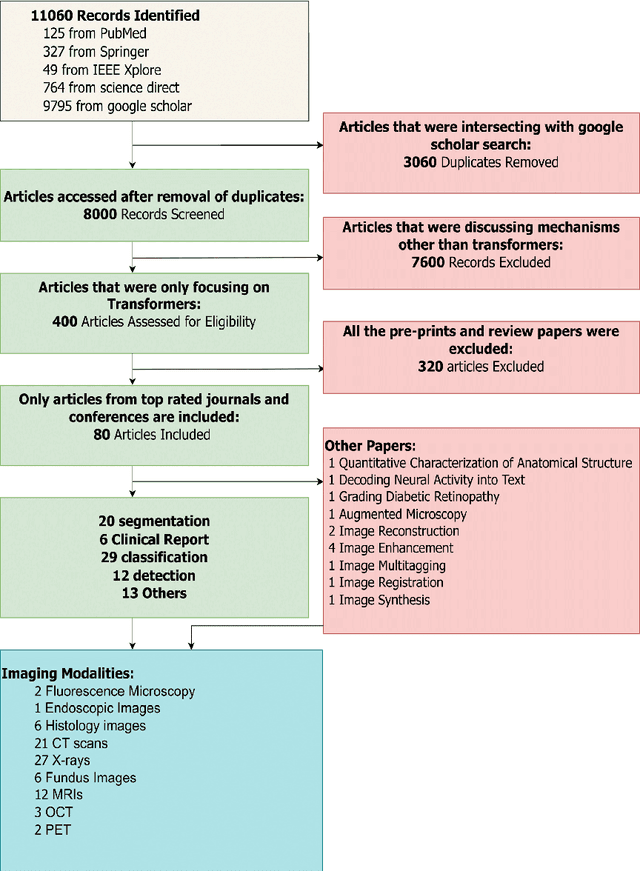
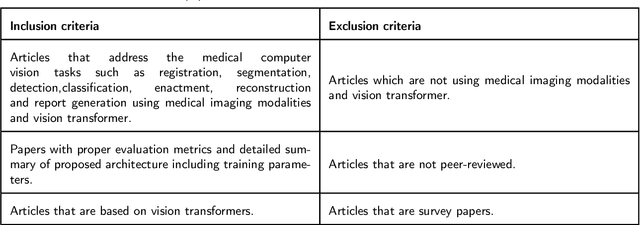
Abstract:Recent escalation in the field of computer vision underpins a huddle of algorithms with the magnificent potential to unravel the information contained within images. These computer vision algorithms are being practised in medical image analysis and are transfiguring the perception and interpretation of Imaging data. Among these algorithms, Vision Transformers are evolved as one of the most contemporary and dominant architectures that are being used in the field of computer vision. These are immensely utilized by a plenty of researchers to perform new as well as former experiments. Here, in this article we investigate the intersection of Vision Transformers and Medical images and proffered an overview of various ViTs based frameworks that are being used by different researchers in order to decipher the obstacles in Medical Computer Vision. We surveyed the application of Vision transformers in different areas of medical computer vision such as image-based disease classification, anatomical structure segmentation, registration, region-based lesion Detection, captioning, report generation, reconstruction using multiple medical imaging modalities that greatly assist in medical diagnosis and hence treatment process. Along with this, we also demystify several imaging modalities used in Medical Computer Vision. Moreover, to get more insight and deeper understanding, self-attention mechanism of transformers is also explained briefly. Conclusively, we also put some light on available data sets, adopted methodology, their performance measures, challenges and their solutions in form of discussion. We hope that this review article will open future directions for researchers in medical computer vision.
Person Re-identification: A Retrospective on Domain Specific Open Challenges and Future Trends
Feb 26, 2022
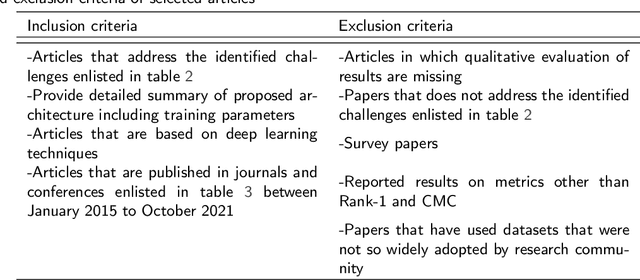
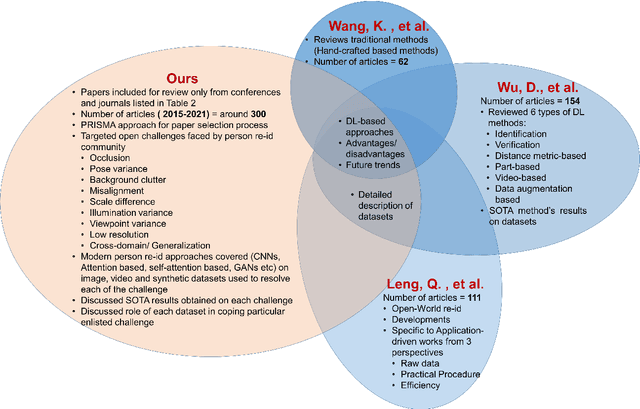
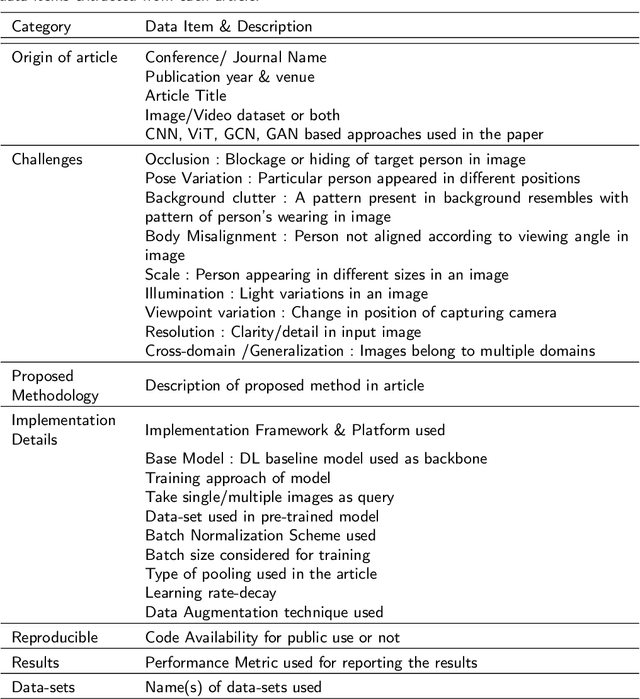
Abstract:Person re-identification (Re-ID) is one of the primary components of an automated visual surveillance system. It aims to automatically identify/search persons in a multi-camera network having non-overlapping field-of-views. Owing to its potential in various applications and research significance, a plethora of deep learning based re-Id approaches have been proposed in the recent years. However, there exist several vision related challenges, e.g., occlusion, pose scale \& viewpoint variance, background clutter, person misalignment and cross-domain generalization across camera modalities, which makes the problem of re-Id still far from being solved. Majority of the proposed approaches directly or indirectly aim to solve one or multiple of these existing challenges. In this context, a comprehensive review of current re-ID approaches in solving theses challenges is needed to analyze and focus on particular aspects for further advancements. At present, such a focused review does not exist and henceforth in this paper, we have presented a systematic challenge-specific literature survey of 230+ papers between the years of 2015-21. For the first time a survey of this type have been presented where the person re-Id approaches are reviewed in such solution-oriented perspective. Moreover, we have presented several diversified prominent developing trends in the respective research domain which will provide a visionary perspective regarding ongoing person re-Id research and eventually help to develop practical real world solutions.
Orientation Aware Weapons Detection In Visual Data : A Benchmark Dataset
Dec 04, 2021
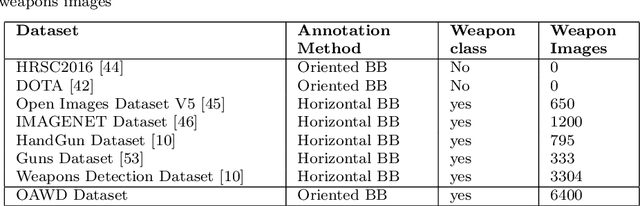


Abstract:Automatic detection of weapons is significant for improving security and well being of individuals, nonetheless, it is a difficult task due to large variety of size, shape and appearance of weapons. View point variations and occlusion also are reasons which makes this task more difficult. Further, the current object detection algorithms process rectangular areas, however a slender and long rifle may really cover just a little portion of area and the rest may contain unessential details. To overcome these problem, we propose a CNN architecture for Orientation Aware Weapons Detection, which provides oriented bounding box with improved weapons detection performance. The proposed model provides orientation not only using angle as classification problem by dividing angle into eight classes but also angle as regression problem. For training our model for weapon detection a new dataset comprising of total 6400 weapons images is gathered from the web and then manually annotated with position oriented bounding boxes. Our dataset provides not only oriented bounding box as ground truth but also horizontal bounding box. We also provide our dataset in multiple formats of modern object detectors for further research in this area. The proposed model is evaluated on this dataset, and the comparative analysis with off-the shelf object detectors yields superior performance of proposed model, measured with standard evaluation strategies. The dataset and the model implementation are made publicly available at this link: https://bit.ly/2TyZICF.
Context-Aware Convolutional Neural Network for Grading of Colorectal Cancer Histology Images
Jul 22, 2019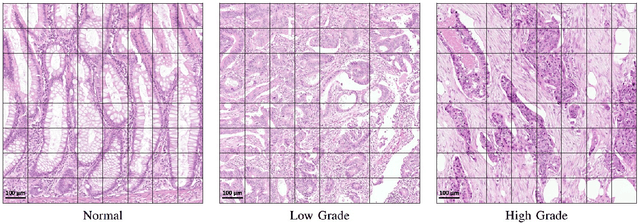
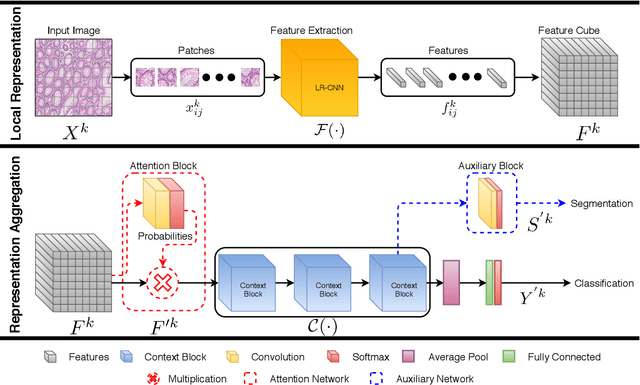
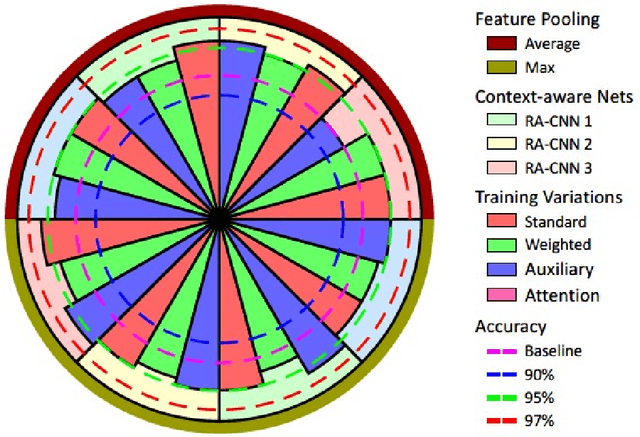

Abstract:Digital histology images are amenable to the application of convolutional neural network (CNN) for analysis due to the sheer size of pixel data present in them. CNNs are generally used for representation learning from small image patches (e.g. 224x224) extracted from digital histology images due to computational and memory constraints. However, this approach does not incorporate high-resolution contextual information in histology images. We propose a novel way to incorporate larger context by a context-aware neural network based on images with a dimension of 1,792x1,792 pixels. The proposed framework first encodes the local representation of a histology image into high dimensional features then aggregates the features by considering their spatial organization to make a final prediction. The proposed method is evaluated for colorectal cancer grading and breast cancer classification. A comprehensive analysis of some variants of the proposed method is presented. Our method outperformed the traditional patch-based approaches, problem-specific methods, and existing context-based methods quantitatively by a margin of 3.61%. Code and dataset related information is available at this link: https://tia-lab.github.io/Context-Aware-CNN
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge