Muhammad Anwaar Khalid
'Aariz: A Benchmark Dataset for Automatic Cephalometric Landmark Detection and CVM Stage Classification
Feb 15, 2023



Abstract:The accurate identification and precise localization of cephalometric landmarks enable the classification and quantification of anatomical abnormalities. The traditional way of marking cephalometric landmarks on lateral cephalograms is a monotonous and time-consuming job. Endeavours to develop automated landmark detection systems have persistently been made, however, they are inadequate for orthodontic applications due to unavailability of a reliable dataset. We proposed a new state-of-the-art dataset to facilitate the development of robust AI solutions for quantitative morphometric analysis. The dataset includes 1000 lateral cephalometric radiographs (LCRs) obtained from 7 different radiographic imaging devices with varying resolutions, making it the most diverse and comprehensive cephalometric dataset to date. The clinical experts of our team meticulously annotated each radiograph with 29 cephalometric landmarks, including the most significant soft tissue landmarks ever marked in any publicly available dataset. Additionally, our experts also labelled the cervical vertebral maturation (CVM) stage of the patient in a radiograph, making this dataset the first standard resource for CVM classification. We believe that this dataset will be instrumental in the development of reliable automated landmark detection frameworks for use in orthodontics and beyond.
CEPHA29: Automatic Cephalometric Landmark Detection Challenge 2023
Dec 09, 2022



Abstract:Quantitative cephalometric analysis is the most widely used clinical and research tool in modern orthodontics. Accurate localization of cephalometric landmarks enables the quantification and classification of anatomical abnormalities, however, the traditional manual way of marking these landmarks is a very tedious job. Endeavours have constantly been made to develop automated cephalometric landmark detection systems but they are inadequate for orthodontic applications. The fundamental reason for this is that the amount of publicly available datasets as well as the images provided for training in these datasets are insufficient for an AI model to perform well. To facilitate the development of robust AI solutions for morphometric analysis, we organise the CEPHA29 Automatic Cephalometric Landmark Detection Challenge in conjunction with IEEE International Symposium on Biomedical Imaging (ISBI 2023). In this context, we provide the largest known publicly available dataset, consisting of 1000 cephalometric X-ray images. We hope that our challenge will not only derive forward research and innovation in automatic cephalometric landmark identification but will also signal the beginning of a new era in the discipline.
Vision Transformers in Medical Computer Vision -- A Contemplative Retrospection
Mar 29, 2022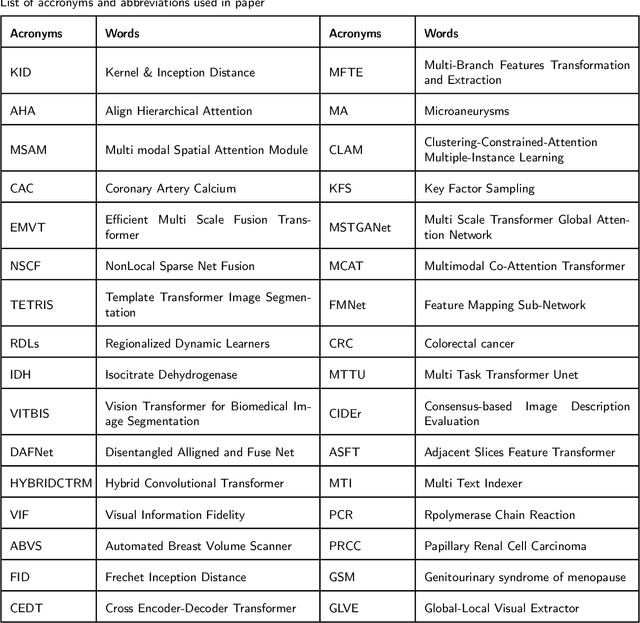
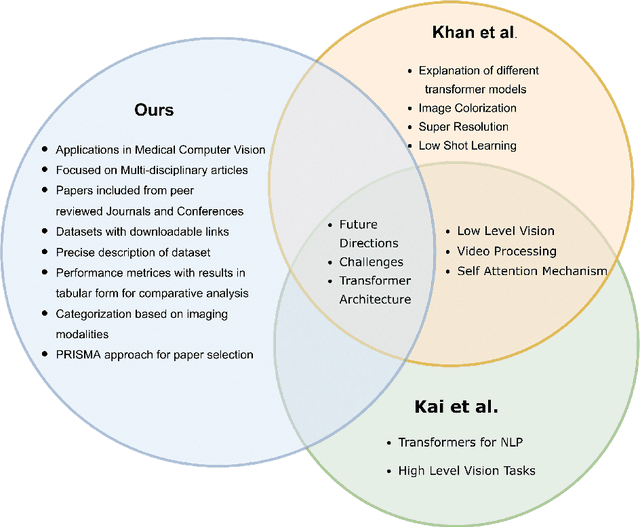
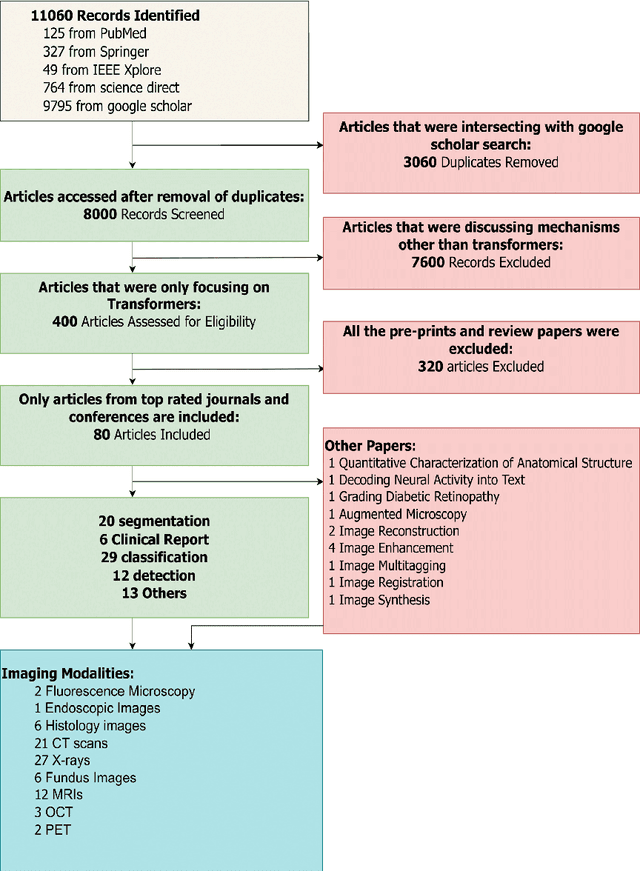
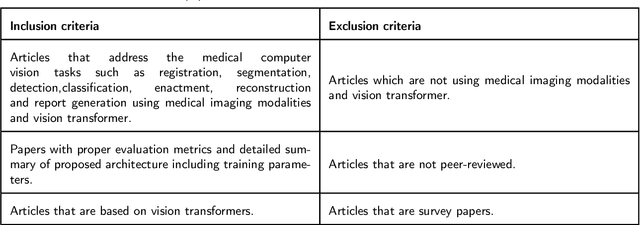
Abstract:Recent escalation in the field of computer vision underpins a huddle of algorithms with the magnificent potential to unravel the information contained within images. These computer vision algorithms are being practised in medical image analysis and are transfiguring the perception and interpretation of Imaging data. Among these algorithms, Vision Transformers are evolved as one of the most contemporary and dominant architectures that are being used in the field of computer vision. These are immensely utilized by a plenty of researchers to perform new as well as former experiments. Here, in this article we investigate the intersection of Vision Transformers and Medical images and proffered an overview of various ViTs based frameworks that are being used by different researchers in order to decipher the obstacles in Medical Computer Vision. We surveyed the application of Vision transformers in different areas of medical computer vision such as image-based disease classification, anatomical structure segmentation, registration, region-based lesion Detection, captioning, report generation, reconstruction using multiple medical imaging modalities that greatly assist in medical diagnosis and hence treatment process. Along with this, we also demystify several imaging modalities used in Medical Computer Vision. Moreover, to get more insight and deeper understanding, self-attention mechanism of transformers is also explained briefly. Conclusively, we also put some light on available data sets, adopted methodology, their performance measures, challenges and their solutions in form of discussion. We hope that this review article will open future directions for researchers in medical computer vision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge