Moran Rubin
Knowing the Distance: Understanding the Gap Between Synthetic and Real Data For Face Parsing
Mar 27, 2023
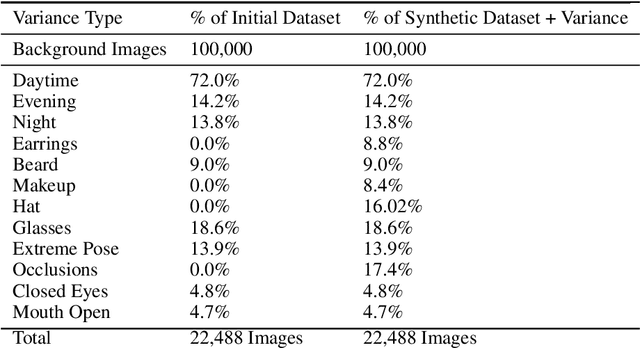

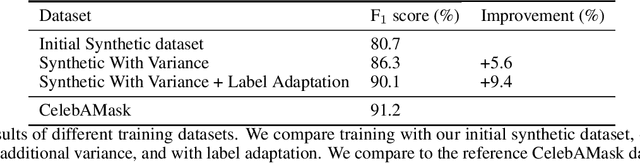
Abstract:The use of synthetic data for training computer vision algorithms has become increasingly popular due to its cost-effectiveness, scalability, and ability to provide accurate multi-modality labels. Although recent studies have demonstrated impressive results when training networks solely on synthetic data, there remains a performance gap between synthetic and real data that is commonly attributed to lack of photorealism. The aim of this study is to investigate the gap in greater detail for the face parsing task. We differentiate between three types of gaps: distribution gap, label gap, and photorealism gap. Our findings show that the distribution gap is the largest contributor to the performance gap, accounting for over 50% of the gap. By addressing this gap and accounting for the labels gap, we demonstrate that a model trained on synthetic data achieves comparable results to one trained on a similar amount of real data. This suggests that synthetic data is a viable alternative to real data, especially when real data is limited or difficult to obtain. Our study highlights the importance of content diversity in synthetic datasets and challenges the notion that the photorealism gap is the most critical factor affecting the performance of computer vision models trained on synthetic data.
TOP-GAN: Label-Free Cancer Cell Classification Using Deep Learning with a Small Training Set
Dec 17, 2018



Abstract:We propose a new deep learning approach for medical imaging that copes with the problem of a small training set, the main bottleneck of deep learning, and apply it for classification of healthy and cancer cells acquired by quantitative phase imaging. The proposed method, called transferring of pre-trained generative adversarial network (TOP-GAN), is a hybridization between transfer learning and generative adversarial networks (GANs). Healthy cells and cancer cells of different metastatic potential have been imaged by low-coherence off-axis holography. After the acquisition, the optical path delay maps of the cells have been extracted and directly used as an input to the deep networks. In order to cope with the small number of classified images, we have used GANs to train a large number of unclassified images from another cell type (sperm cells). After this preliminary training, and after transforming the last layer of the network with new ones, we have designed an automatic classifier for the correct cell type (healthy/primary cancer/metastatic cancer) with 90-99% accuracy, although small training sets of down to several images have been used. These results are better in comparison to other classic methods that aim at coping with the same problem of a small training set. We believe that our approach makes the combination of holographic microscopy and deep learning networks more accessible to the medical field by enabling a rapid, automatic and accurate classification in stain-free imaging flow cytometry. Furthermore, our approach is expected to be applicable to many other medical image classification tasks, suffering from a small training set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge