Miron B. Kursa
Generalised Random Forest Space Overview
Jan 17, 2015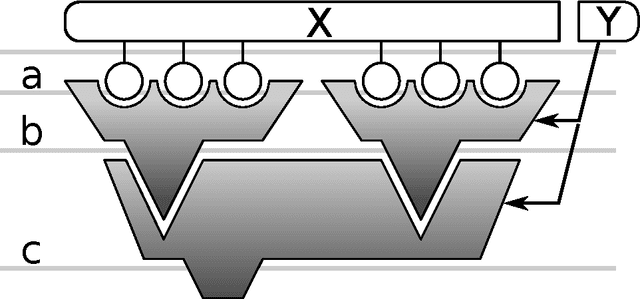
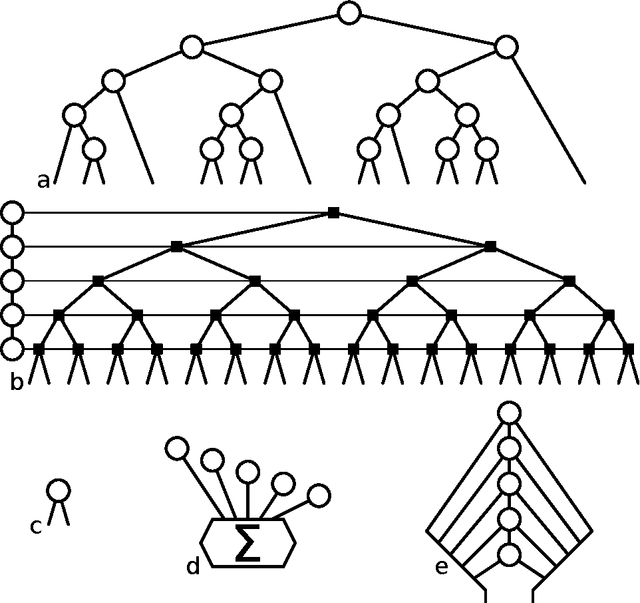
Abstract:Assuming a view of the Random Forest as a special case of a nested ensemble of interchangeable modules, we construct a generalisation space allowing one to easily develop novel methods based on this algorithm. We discuss the role and required properties of modules at each level, especially in context of some already proposed RF generalisations.
rFerns: An Implementation of the Random Ferns Method for General-Purpose Machine Learning
Nov 14, 2014



Abstract:In this paper I present an extended implementation of the Random ferns algorithm contained in the R package rFerns. It differs from the original by the ability of consuming categorical and numerical attributes instead of only binary ones. Also, instead of using simple attribute subspace ensemble it employs bagging and thus produce error approximation and variable importance measure modelled after Random forest algorithm. I also present benchmarks' results which show that although Random ferns' accuracy is mostly smaller than achieved by Random forest, its speed and good quality of importance measure it provides make rFerns a reasonable choice for a specific applications.
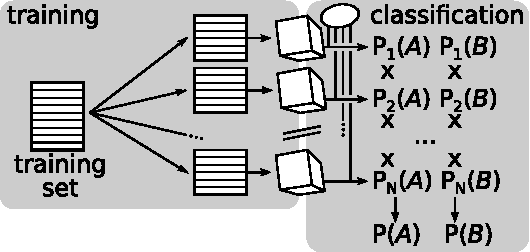
Multi-label Ferns for Efficient Recognition of Musical Instruments in Recordings
Mar 30, 2014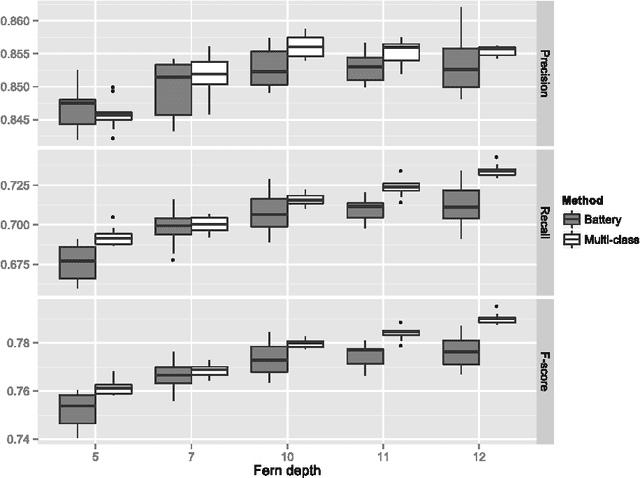
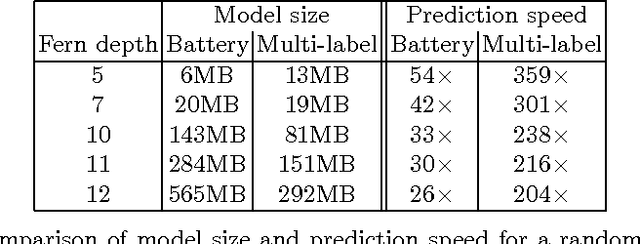
Abstract:In this paper we introduce multi-label ferns, and apply this technique for automatic classification of musical instruments in audio recordings. We compare the performance of our proposed method to a set of binary random ferns, using jazz recordings as input data. Our main result is obtaining much faster classification and higher F-score. We also achieve substantial reduction of the model size.
Robustness of Random Forest-based gene selection methods
Oct 18, 2013



Abstract:Gene selection is an important part of microarray data analysis because it provides information that can lead to a better mechanistic understanding of an investigated phenomenon. At the same time, gene selection is very difficult because of the noisy nature of microarray data. As a consequence, gene selection is often performed with machine learning methods. The Random Forest method is particularly well suited for this purpose. In this work, four state-of-the-art Random Forest-based feature selection methods were compared in a gene selection context. The analysis focused on the stability of selection because, although it is necessary for determining the significance of results, it is often ignored in similar studies. The comparison of post-selection accuracy in the validation of Random Forest classifiers revealed that all investigated methods were equivalent in this context. However, the methods substantially differed with respect to the number of selected genes and the stability of selection. Of the analysed methods, the Boruta algorithm predicted the most genes as potentially important. The post-selection classifier error rate, which is a frequently used measure, was found to be a potentially deceptive measure of gene selection quality. When the number of consistently selected genes was considered, the Boruta algorithm was clearly the best. Although it was also the most computationally intensive method, the Boruta algorithm's computational demands could be reduced to levels comparable to those of other algorithms by replacing the Random Forest importance with a comparable measure from Random Ferns (a similar but simplified classifier). Despite their design assumptions, the minimal optimal selection methods, were found to select a high fraction of false positives.
A Comparison of Random Forests and Ferns on Recognition of Instruments in Jazz Recordings
May 22, 2013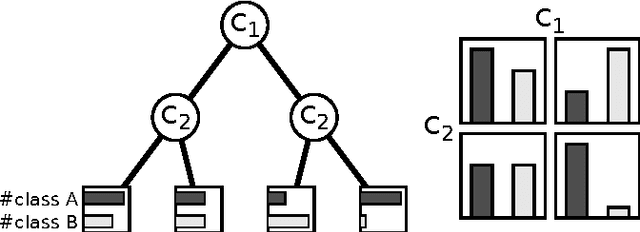


Abstract:In this paper, we first apply random ferns for classification of real music recordings of a jazz band. No initial segmentation of audio data is assumed, i.e., no onset, offset, nor pitch data are needed. The notion of random ferns is described in the paper, to familiarize the reader with this classification algorithm, which was introduced quite recently and applied so far in image recognition tasks. The performance of random ferns is compared with random forests for the same data. The results of experiments are presented in the paper, and conclusions are drawn.
The All Relevant Feature Selection using Random Forest
Jun 25, 2011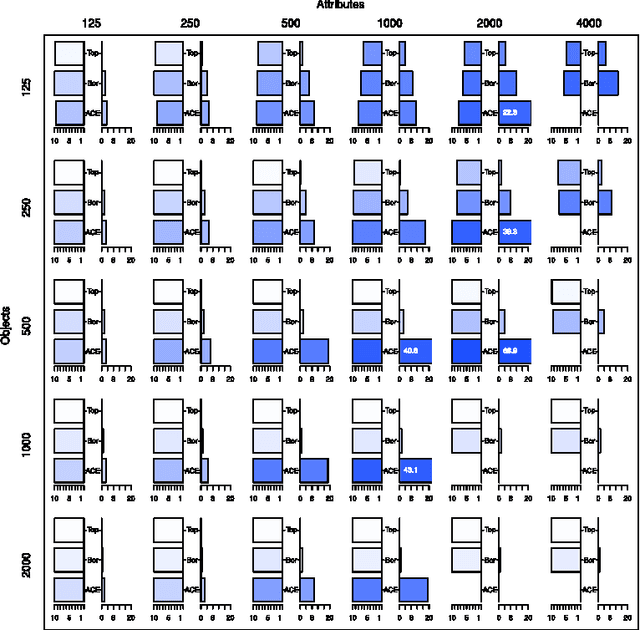
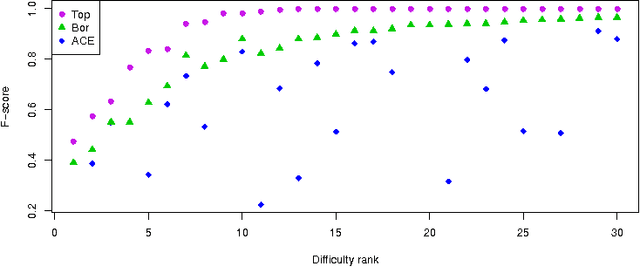
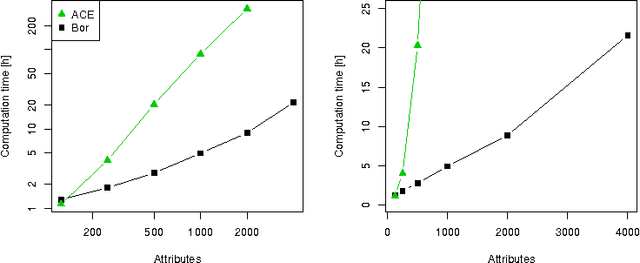
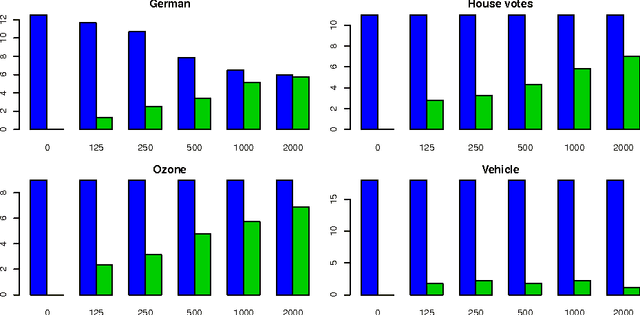
Abstract:In this paper we examine the application of the random forest classifier for the all relevant feature selection problem. To this end we first examine two recently proposed all relevant feature selection algorithms, both being a random forest wrappers, on a series of synthetic data sets with varying size. We show that reasonable accuracy of predictions can be achieved and that heuristic algorithms that were designed to handle the all relevant problem, have performance that is close to that of the reference ideal algorithm. Then, we apply one of the algorithms to four families of semi-synthetic data sets to assess how the properties of particular data set influence results of feature selection. Finally we test the procedure using a well-known gene expression data set. The relevance of nearly all previously established important genes was confirmed, moreover the relevance of several new ones is discovered.
Random forest models of the retention constants in the thin layer chromatography
Jun 16, 2011



Abstract:In the current study we examine an application of the machine learning methods to model the retention constants in the thin layer chromatography (TLC). This problem can be described with hundreds or even thousands of descriptors relevant to various molecular properties, most of them redundant and not relevant for the retention constant prediction. Hence we employed feature selection to significantly reduce the number of attributes. Additionally we have tested application of the bagging procedure to the feature selection. The random forest regression models were built using selected variables. The resulting models have better correlation with the experimental data than the reference models obtained with linear regression. The cross-validation confirms robustness of the models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge