Michael Shaw
Autonomous object harvesting using synchronized optoelectronic microrobots
Mar 08, 2021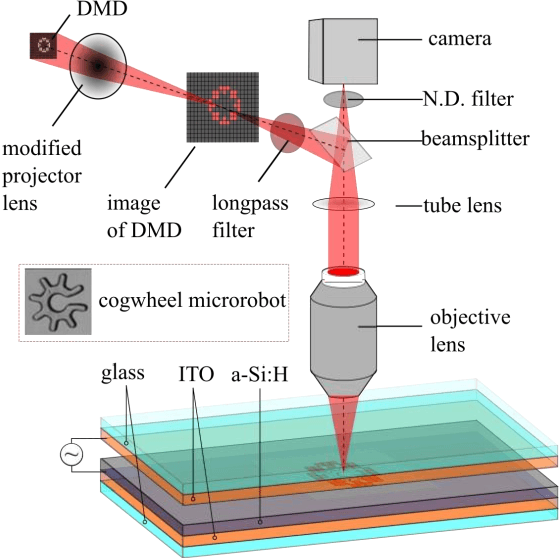
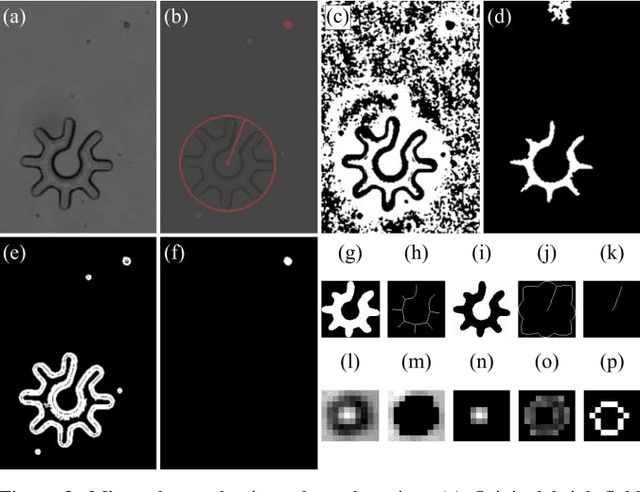
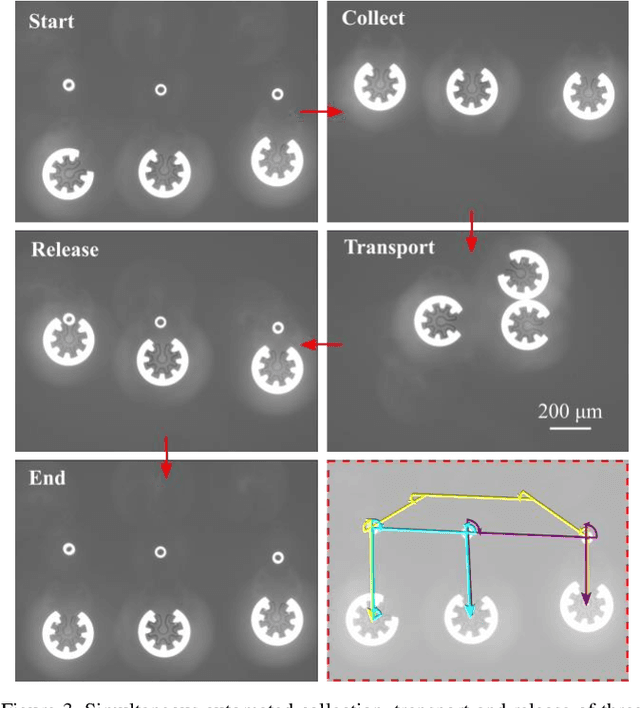
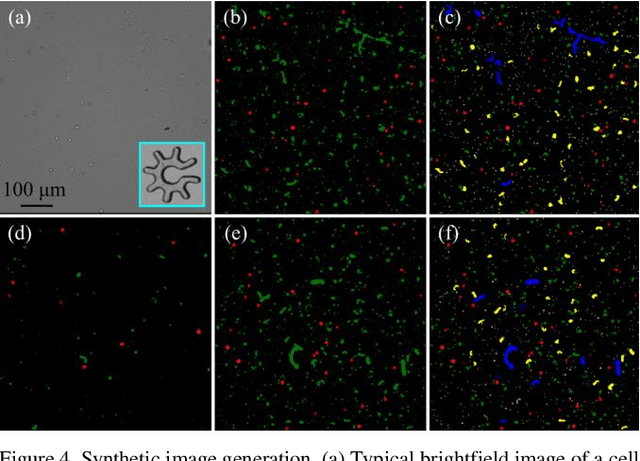
Abstract:Optoelectronic tweezer-driven microrobots (OETdMs) are a versatile micromanipulation technology based on the use of light induced dielectrophoresis to move small dielectric structures (microrobots) across a photoconductive substrate. The microrobots in turn can be used to exert forces on secondary objects and carry out a wide range of micromanipulation operations, including collecting, transporting and depositing microscopic cargos. In contrast to alternative (direct) micromanipulation techniques, OETdMs are relatively gentle, making them particularly well suited to interacting with sensitive objects such as biological cells. However, at present such systems are used exclusively under manual control by a human operator. This limits the capacity for simultaneous control of multiple microrobots, reducing both experimental throughput and the possibility of cooperative multi-robot operations. In this article, we describe an approach to automated targeting and path planning to enable open-loop control of multiple microrobots. We demonstrate the performance of the method in practice, using microrobots to simultaneously collect, transport and deposit silica microspheres. Using computational simulations based on real microscopic image data, we investigate the capacity of microrobots to collect target cells from within a dissociated tissue culture. Our results indicate the feasibility of using OETdMs to autonomously carry out micromanipulation tasks within complex, unstructured environments.
Whole-Sample Mapping of Cancerous and Benign Tissue Properties
Jul 23, 2019



Abstract:Structural and mechanical differences between cancerous and healthy tissue give rise to variations in macroscopic properties such as visual appearance and elastic modulus that show promise as signatures for early cancer detection. Atomic force microscopy (AFM) has been used to measure significant differences in stiffness between cancerous and healthy cells owing to its high force sensitivity and spatial resolution, however due to absorption and scattering of light, it is often challenging to accurately locate where AFM measurements have been made on a bulk tissue sample. In this paper we describe an image registration method that localizes AFM elastic stiffness measurements with high-resolution images of haematoxylin and eosin (H\&E)-stained tissue to within 1.5 microns. Color RGB images are segmented into three structure types (lumen, cells and stroma) by a neural network classifier trained on ground-truth pixel data obtained through k-means clustering in HSV color space. Using the localized stiffness maps and corresponding structural information, a whole-sample stiffness map is generated with a region matching and interpolation algorithm that associates similar structures with measured stiffness values. We present results showing significant differences in stiffness between healthy and cancerous liver tissue and discuss potential applications of this technique.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge