Mehmet Kurt
Clinical Interpretability of Deep Learning Segmentation Through Shapley-Derived Agreement and Uncertainty Metrics
Dec 08, 2025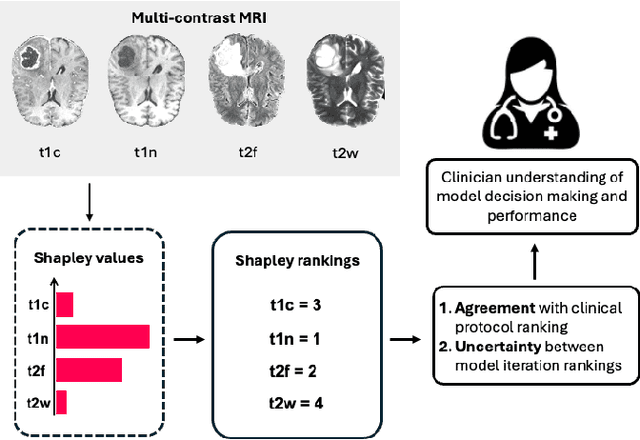
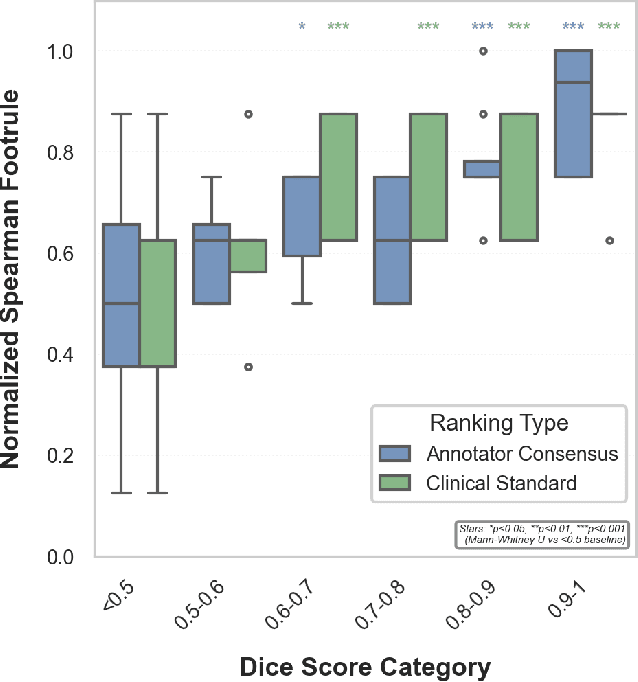
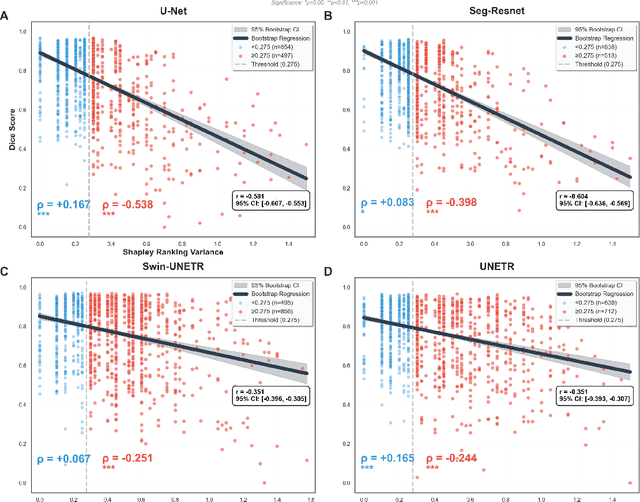
Abstract:Segmentation is the identification of anatomical regions of interest, such as organs, tissue, and lesions, serving as a fundamental task in computer-aided diagnosis in medical imaging. Although deep learning models have achieved remarkable performance in medical image segmentation, the need for explainability remains critical for ensuring their acceptance and integration in clinical practice, despite the growing research attention in this area. Our approach explored the use of contrast-level Shapley values, a systematic perturbation of model inputs to assess feature importance. While other studies have investigated gradient-based techniques through identifying influential regions in imaging inputs, Shapley values offer a broader, clinically aligned approach, explaining how model performance is fairly attributed to certain imaging contrasts over others. Using the BraTS 2024 dataset, we generated rankings for Shapley values for four MRI contrasts across four model architectures. Two metrics were proposed from the Shapley ranking: agreement between model and ``clinician" imaging ranking, and uncertainty quantified through Shapley ranking variance across cross-validation folds. Higher-performing cases (Dice \textgreater0.6) showed significantly greater agreement with clinical rankings. Increased Shapley ranking variance correlated with decreased performance (U-Net: $r=-0.581$). These metrics provide clinically interpretable proxies for model reliability, helping clinicians better understand state-of-the-art segmentation models.
How We Won the ISLES'24 Challenge by Preprocessing
May 23, 2025Abstract:Stroke is among the top three causes of death worldwide, and accurate identification of stroke lesion boundaries is critical for diagnosis and treatment. Supervised deep learning methods have emerged as the leading solution for stroke lesion segmentation but require large, diverse, and annotated datasets. The ISLES'24 challenge addresses this need by providing longitudinal stroke imaging data, including CT scans taken on arrival to the hospital and follow-up MRI taken 2-9 days from initial arrival, with annotations derived from follow-up MRI. Importantly, models submitted to the ISLES'24 challenge are evaluated using only CT inputs, requiring prediction of lesion progression that may not be visible in CT scans for segmentation. Our winning solution shows that a carefully designed preprocessing pipeline including deep-learning-based skull stripping and custom intensity windowing is beneficial for accurate segmentation. Combined with a standard large residual nnU-Net architecture for segmentation, this approach achieves a mean test Dice of 28.5 with a standard deviation of 21.27.
Here Comes the Explanation: A Shapley Perspective on Multi-contrast Medical Image Segmentation
Apr 06, 2025Abstract:Deep learning has been successfully applied to medical image segmentation, enabling accurate identification of regions of interest such as organs and lesions. This approach works effectively across diverse datasets, including those with single-image contrast, multi-contrast, and multimodal imaging data. To improve human understanding of these black-box models, there is a growing need for Explainable AI (XAI) techniques for model transparency and accountability. Previous research has primarily focused on post hoc pixel-level explanations, using methods gradient-based and perturbation-based apporaches. These methods rely on gradients or perturbations to explain model predictions. However, these pixel-level explanations often struggle with the complexity inherent in multi-contrast magnetic resonance imaging (MRI) segmentation tasks, and the sparsely distributed explanations have limited clinical relevance. In this study, we propose using contrast-level Shapley values to explain state-of-the-art models trained on standard metrics used in brain tumor segmentation. Our results demonstrate that Shapley analysis provides valuable insights into different models' behavior used for tumor segmentation. We demonstrated a bias for U-Net towards over-weighing T1-contrast and FLAIR, while Swin-UNETR provided a cross-contrast understanding with balanced Shapley distribution.
An Ensemble Approach for Brain Tumor Segmentation and Synthesis
Nov 26, 2024Abstract:The integration of machine learning in magnetic resonance imaging (MRI), specifically in neuroimaging, is proving to be incredibly effective, leading to better diagnostic accuracy, accelerated image analysis, and data-driven insights, which can potentially transform patient care. Deep learning models utilize multiple layers of processing to capture intricate details of complex data, which can then be used on a variety of tasks, including brain tumor classification, segmentation, image synthesis, and registration. Previous research demonstrates high accuracy in tumor segmentation using various model architectures, including nn-UNet and Swin-UNet. U-Mamba, which uses state space modeling, also achieves high accuracy in medical image segmentation. To leverage these models, we propose a deep learning framework that ensembles these state-of-the-art architectures to achieve accurate segmentation and produce finely synthesized images.
Re-DiffiNet: Modeling discrepancies loss in tumor segmentation using diffusion models
Feb 15, 2024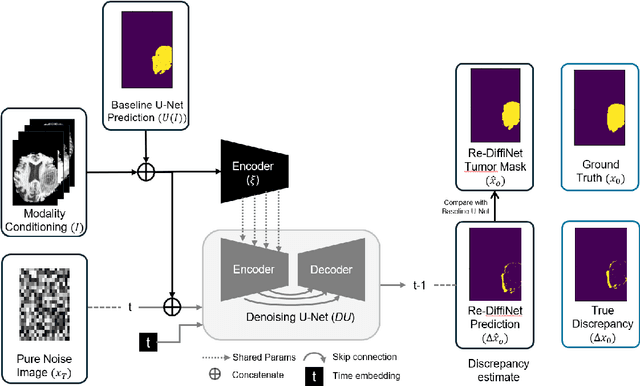
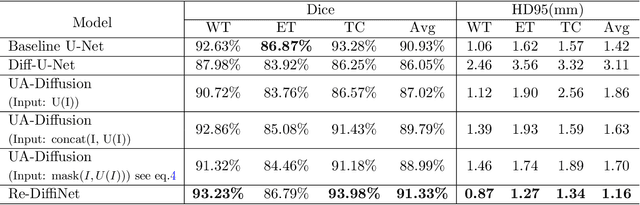
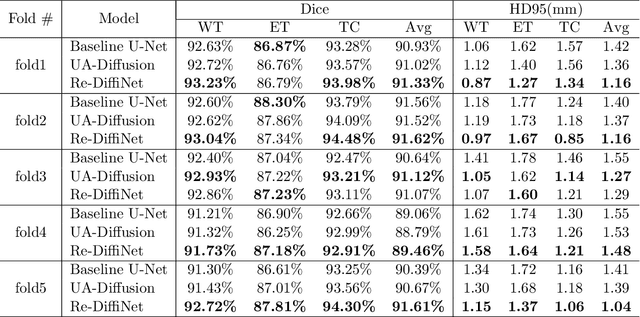
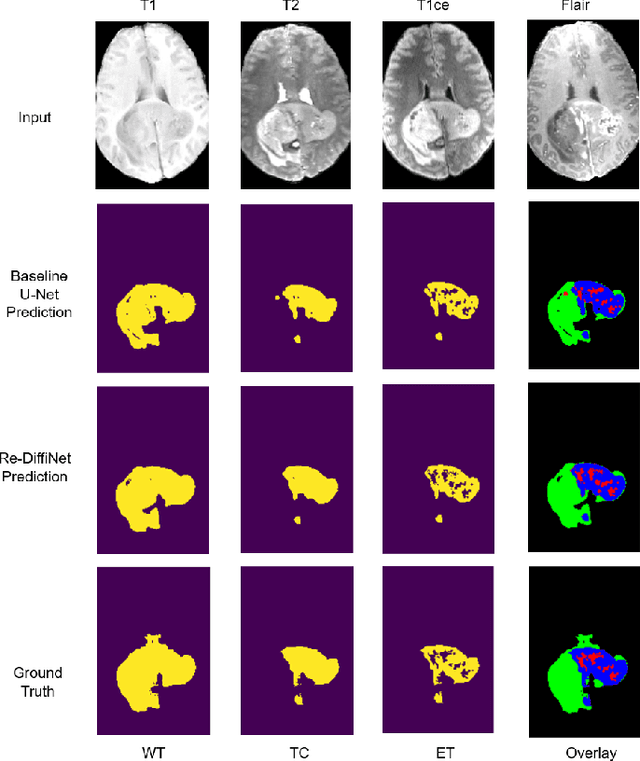
Abstract:Identification of tumor margins is essential for surgical decision-making for glioblastoma patients and provides reliable assistance for neurosurgeons. Despite improvements in deep learning architectures for tumor segmentation over the years, creating a fully autonomous system suitable for clinical floors remains a formidable challenge because the model predictions have not yet reached the desired level of accuracy and generalizability for clinical applications. Generative modeling techniques have seen significant improvements in recent times. Specifically, Generative Adversarial Networks (GANs) and Denoising-diffusion-based models (DDPMs) have been used to generate higher-quality images with fewer artifacts and finer attributes. In this work, we introduce a framework called Re-Diffinet for modeling the discrepancy between the outputs of a segmentation model like U-Net and the ground truth, using DDPMs. By explicitly modeling the discrepancy, the results show an average improvement of 0.55\% in the Dice score and 16.28\% in HD95 from cross-validation over 5-folds, compared to the state-of-the-art U-Net segmentation model.
An Optimization Framework for Processing and Transfer Learning for the Brain Tumor Segmentation
Feb 10, 2024



Abstract:Tumor segmentation from multi-modal brain MRI images is a challenging task due to the limited samples, high variance in shapes and uneven distribution of tumor morphology. The performance of automated medical image segmentation has been significant improvement by the recent advances in deep learning. However, the model predictions have not yet reached the desired level for clinical use in terms of accuracy and generalizability. In order to address the distinct problems presented in Challenges 1, 2, and 3 of BraTS 2023, we have constructed an optimization framework based on a 3D U-Net model for brain tumor segmentation. This framework incorporates a range of techniques, including various pre-processing and post-processing techniques, and transfer learning. On the validation datasets, this multi-modality brain tumor segmentation framework achieves an average lesion-wise Dice score of 0.79, 0.72, 0.74 on Challenges 1, 2, 3 respectively.
3D Inception-Based TransMorph: Pre- and Post-operative Multi-contrast MRI Registration in Brain Tumors
Dec 08, 2022Abstract:Deformable image registration is a key task in medical image analysis. The Brain Tumor Sequence Registration challenge (BraTS-Reg) aims at establishing correspondences between pre-operative and follow-up scans of the same patient diagnosed with an adult brain diffuse high-grade glioma and intends to address the challenging task of registering longitudinal data with major tissue appearance changes. In this work, we proposed a two-stage cascaded network based on the Inception and TransMorph models. The dataset for each patient was comprised of a native pre-contrast (T1), a contrast-enhanced T1-weighted (T1-CE), a T2-weighted (T2), and a Fluid Attenuated Inversion Recovery (FLAIR). The Inception model was used to fuse the 4 image modalities together and extract the most relevant information. Then, a variant of the TransMorph architecture was adapted to generate the displacement fields. The Loss function was composed of a standard image similarity measure, a diffusion regularizer, and an edge-map similarity measure added to overcome intensity dependence and reinforce correct boundary deformation. We observed that the addition of the Inception module substantially increased the performance of the network. Additionally, performing an initial affine registration before training the model showed improved accuracy in the landmark error measurements between pre and post-operative MRIs. We observed that our best model composed of the Inception and TransMorph architectures while using an initially affine registered dataset had the best performance with a median absolute error of 2.91 (initial error = 7.8). We achieved 6th place at the time of model submission in the final testing phase of the BraTS-Reg challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge