Md. Tasnim Jawad
Acute Lymphoblastic Leukemia Detection from Microscopic Images Using Weighted Ensemble of Convolutional Neural Networks
May 09, 2021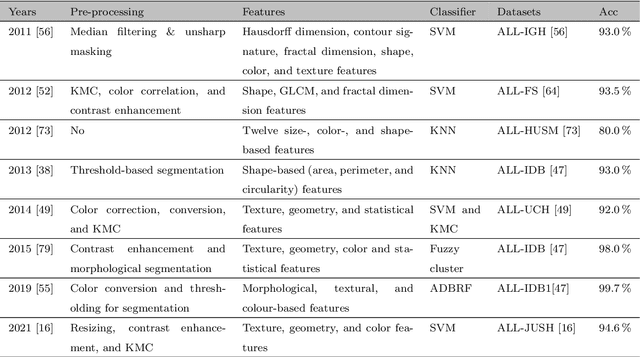
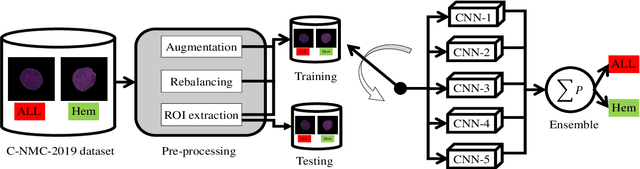
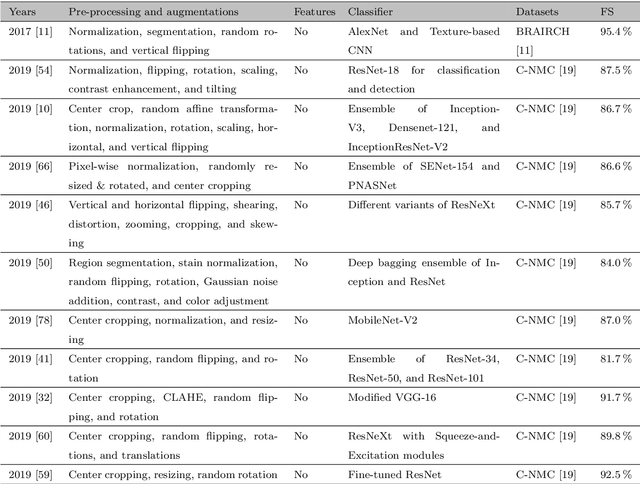
Abstract:Acute Lymphoblastic Leukemia (ALL) is a blood cell cancer characterized by numerous immature lymphocytes. Even though automation in ALL prognosis is an essential aspect of cancer diagnosis, it is challenging due to the morphological correlation between malignant and normal cells. The traditional ALL classification strategy demands experienced pathologists to carefully read the cell images, which is arduous, time-consuming, and often suffers inter-observer variations. This article has automated the ALL detection task from microscopic cell images, employing deep Convolutional Neural Networks (CNNs). We explore the weighted ensemble of different deep CNNs to recommend a better ALL cell classifier. The weights for the ensemble candidate models are estimated from their corresponding metrics, such as accuracy, F1-score, AUC, and kappa values. Various data augmentations and pre-processing are incorporated for achieving a better generalization of the network. We utilize the publicly available C-NMC-2019 ALL dataset to conduct all the comprehensive experiments. Our proposed weighted ensemble model, using the kappa values of the ensemble candidates as their weights, has outputted a weighted F1-score of 88.6 %, a balanced accuracy of 86.2 %, and an AUC of 0.941 in the preliminary test set. The qualitative results displaying the gradient class activation maps confirm that the introduced model has a concentrated learned region. In contrast, the ensemble candidate models, such as Xception, VGG-16, DenseNet-121, MobileNet, and InceptionResNet-V2, separately produce coarse and scatter learned areas for most example cases. Since the proposed kappa value-based weighted ensemble yields a better result for the aimed task in this article, it can experiment in other domains of medical diagnostic applications.
Dermo-DOCTOR: A framework for concurrent skin lesion detection and recognition using a deep convolutional neural network with end-to-end dual encoders
Feb 23, 2021
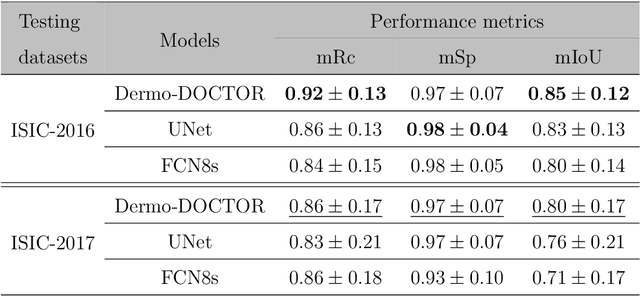
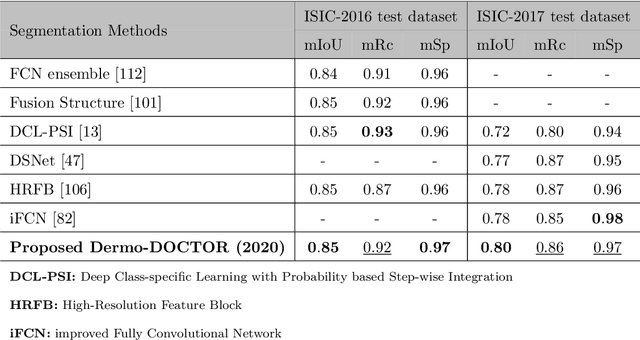
Abstract:Automated skin lesion analysis for simultaneous detection and recognition is still challenging for inter-class homogeneity and intra-class heterogeneity, leading to low generic capability of a Single Convolutional Neural Network (CNN) with limited datasets. This article proposes an end-to-end deep CNN-based framework for simultaneous detection and recognition of the skin lesions, named Dermo-DOCTOR, consisting of two encoders. The feature maps from two encoders are fused channel-wise, called Fused Feature Map (FFM). The FFM is utilized for decoding in the detection sub-network, concatenating each stage of two encoders' outputs with corresponding decoder layers to retrieve the lost spatial information due to pooling in the encoders. For the recognition sub-network, the outputs of three fully connected layers, utilizing feature maps of two encoders and FFM, are aggregated to obtain a final lesion class. We train and evaluate the proposed Dermo-Doctor utilizing two publicly available benchmark datasets, such as ISIC-2016 and ISIC-2017. The achieved segmentation results exhibit mean intersection over unions of 85.0 % and 80.0 % respectively for ISIC-2016 and ISIC-2017 test datasets. The proposed Dermo-DOCTOR also demonstrates praiseworthy success in lesion recognition, providing the areas under the receiver operating characteristic curves of 0.98 and 0.91 respectively for those two datasets. The experimental results show that the proposed Dermo-DOCTOR outperforms the alternative methods mentioned in the literature, designed for skin lesion detection and recognition. As the Dermo-DOCTOR provides better-results on two different test datasets, even with limited training data, it can be an auspicious computer-aided assistive tool for dermatologists.
COVID-19 identification from volumetric chest CT scans using a progressively resized 3D-CNN incorporating segmentation, augmentation, and class-rebalancing
Feb 11, 2021



Abstract:The novel COVID-19 is a global pandemic disease overgrowing worldwide. Computer-aided screening tools with greater sensitivity is imperative for disease diagnosis and prognosis as early as possible. It also can be a helpful tool in triage for testing and clinical supervision of COVID-19 patients. However, designing such an automated tool from non-invasive radiographic images is challenging as many manually annotated datasets are not publicly available yet, which is the essential core requirement of supervised learning schemes. This article proposes a 3D Convolutional Neural Network (CNN)-based classification approach considering both the inter- and intra-slice spatial voxel information. The proposed system is trained in an end-to-end manner on the 3D patches from the whole volumetric CT images to enlarge the number of training samples, performing the ablation studies on patch size determination. We integrate progressive resizing, segmentation, augmentations, and class-rebalancing to our 3D network. The segmentation is a critical prerequisite step for COVID-19 diagnosis enabling the classifier to learn prominent lung features while excluding the outer lung regions of the CT scans. We evaluate all the extensive experiments on a publicly available dataset, named MosMed, having binary- and multi-class chest CT image partitions. Our experimental results are very encouraging, yielding areas under the ROC curve of 0.914 and 0.893 for the binary- and multi-class tasks, respectively, applying 5-fold cross-validations. Our method's promising results delegate it as a favorable aiding tool for clinical practitioners and radiologists to assess COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge