Md. Ashraful Alam
Dermo-DOCTOR: A framework for concurrent skin lesion detection and recognition using a deep convolutional neural network with end-to-end dual encoders
Feb 23, 2021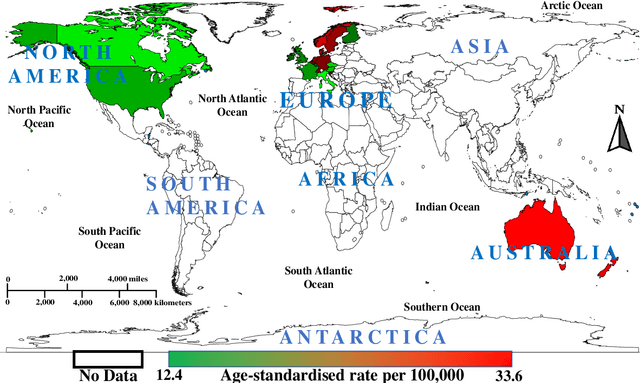
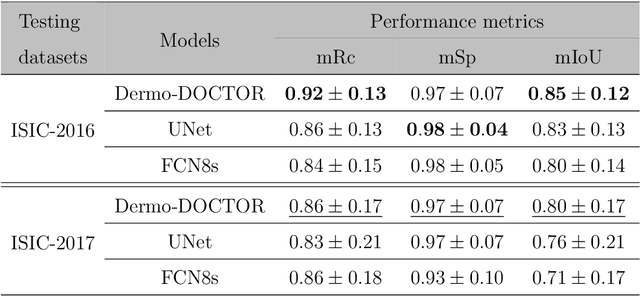
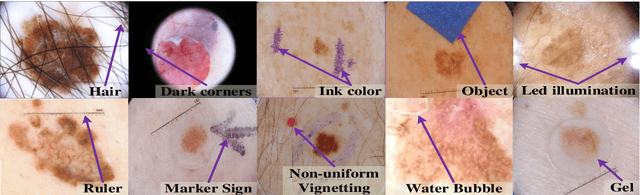
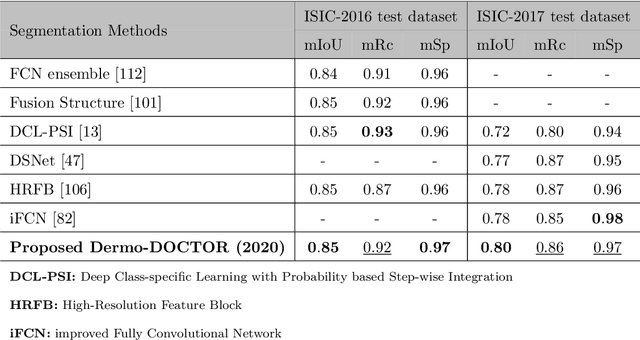
Abstract:Automated skin lesion analysis for simultaneous detection and recognition is still challenging for inter-class homogeneity and intra-class heterogeneity, leading to low generic capability of a Single Convolutional Neural Network (CNN) with limited datasets. This article proposes an end-to-end deep CNN-based framework for simultaneous detection and recognition of the skin lesions, named Dermo-DOCTOR, consisting of two encoders. The feature maps from two encoders are fused channel-wise, called Fused Feature Map (FFM). The FFM is utilized for decoding in the detection sub-network, concatenating each stage of two encoders' outputs with corresponding decoder layers to retrieve the lost spatial information due to pooling in the encoders. For the recognition sub-network, the outputs of three fully connected layers, utilizing feature maps of two encoders and FFM, are aggregated to obtain a final lesion class. We train and evaluate the proposed Dermo-Doctor utilizing two publicly available benchmark datasets, such as ISIC-2016 and ISIC-2017. The achieved segmentation results exhibit mean intersection over unions of 85.0 % and 80.0 % respectively for ISIC-2016 and ISIC-2017 test datasets. The proposed Dermo-DOCTOR also demonstrates praiseworthy success in lesion recognition, providing the areas under the receiver operating characteristic curves of 0.98 and 0.91 respectively for those two datasets. The experimental results show that the proposed Dermo-DOCTOR outperforms the alternative methods mentioned in the literature, designed for skin lesion detection and recognition. As the Dermo-DOCTOR provides better-results on two different test datasets, even with limited training data, it can be an auspicious computer-aided assistive tool for dermatologists.
Multi-class probabilistic atlas-based whole heart segmentation method in cardiac CT and MRI
Feb 03, 2021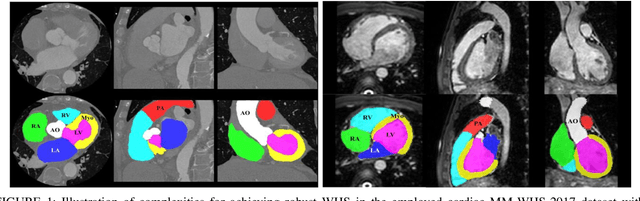
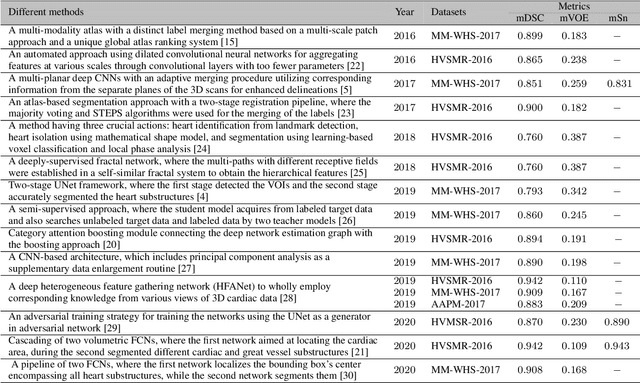
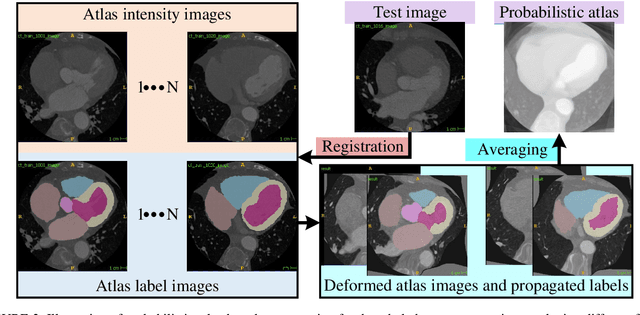
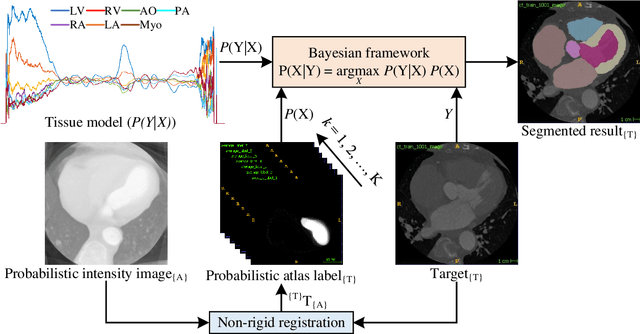
Abstract:Accurate and robust whole heart substructure segmentation is crucial in developing clinical applications, such as computer-aided diagnosis and computer-aided surgery. However, segmentation of different heart substructures is challenging because of inadequate edge or boundary information, the complexity of the background and texture, and the diversity in different substructures' sizes and shapes. This article proposes a framework for multi-class whole heart segmentation employing non-rigid registration-based probabilistic atlas incorporating the Bayesian framework. We also propose a non-rigid registration pipeline utilizing a multi-resolution strategy for obtaining the highest attainable mutual information between the moving and fixed images. We further incorporate non-rigid registration into the expectation-maximization algorithm and implement different deep convolutional neural network-based encoder-decoder networks for ablation studies. All the extensive experiments are conducted utilizing the publicly available dataset for the whole heart segmentation containing 20 MRI and 20 CT cardiac images. The proposed approach exhibits an encouraging achievement, yielding a mean volume overlapping error of 14.5 % for CT scans exceeding the state-of-the-art results by a margin of 1.3 % in terms of the same metric. As the proposed approach provides better-results to delineate the different substructures of the heart, it can be a medical diagnostic aiding tool for helping experts with quicker and more accurate results.
CVR-Net: A deep convolutional neural network for coronavirus recognition from chest radiography images
Jul 21, 2020



Abstract:The novel Coronavirus Disease 2019 (COVID-19) is a global pandemic disease spreading rapidly around the world. A robust and automatic early recognition of COVID-19, via auxiliary computer-aided diagnostic tools, is essential for disease cure and control. The chest radiography images, such as Computed Tomography (CT) and X-ray, and deep Convolutional Neural Networks (CNNs), can be a significant and useful material for designing such tools. However, designing such an automated tool is challenging as a massive number of manually annotated datasets are not publicly available yet, which is the core requirement of supervised learning systems. In this article, we propose a robust CNN-based network, called CVR-Net (Coronavirus Recognition Network), for the automatic recognition of the coronavirus from CT or X-ray images. The proposed end-to-end CVR-Net is a multi-scale-multi-encoder ensemble model, where we have aggregated the outputs from two different encoders and their different scales to obtain the final prediction probability. We train and test the proposed CVR-Net on three different datasets, where the images have collected from different open-source repositories. We compare our proposed CVR-Net with state-of-the-art methods, which are trained and tested on the same datasets. We split three datasets into five different tasks, where each task has a different number of classes, to evaluate the multi-tasking CVR-Net. Our model achieves an overall F1-score & accuracy of 0.997 & 0.998; 0.963 & 0.964; 0.816 & 0.820; 0.961 & 0.961; and 0.780 & 0.780, respectively, for task-1 to task-5. As the CVR-Net provides promising results on the small datasets, it can be an auspicious computer-aided diagnostic tool for the diagnosis of coronavirus to assist the clinical practitioners and radiologists. Our source codes and model are publicly available at https://github.com/kamruleee51/CVR-Net.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge