Maysam Orouskhani
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
Artificial Intelligence in Fetal Resting-State Functional MRI Brain Segmentation: A Comparative Analysis of 3D UNet, VNet, and HighRes-Net Models
Nov 17, 2023Abstract:Introduction: Fetal resting-state functional magnetic resonance imaging (rs-fMRI) is a rapidly evolving field that provides valuable insight into brain development before birth. Accurate segmentation of the fetal brain from the surrounding tissue in nonstationary 3D brain volumes poses a significant challenge in this domain. Current available tools have 0.15 accuracy. Aim: This study introduced a novel application of artificial intelligence (AI) for automated brain segmentation in fetal brain fMRI, magnetic resonance imaging (fMRI). Open datasets were employed to train AI models, assess their performance, and analyze their capabilities and limitations in addressing the specific challenges associated with fetal brain fMRI segmentation. Method: We utilized an open-source fetal functional MRI (fMRI) dataset consisting of 160 cases (reference: fetal-fMRI - OpenNeuro). An AI model for fMRI segmentation was developed using a 5-fold cross-validation methodology. Three AI models were employed: 3D UNet, VNet, and HighResNet. Optuna, an automated hyperparameter-tuning tool, was used to optimize these models. Results and Discussion: The Dice scores of the three AI models (VNet, UNet, and HighRes-net) were compared, including a comparison between manually tuned and automatically tuned models using Optuna. Our findings shed light on the performance of different AI models for fetal resting-state fMRI brain segmentation. Although the VNet model showed promise in this application, further investigation is required to fully explore the potential and limitations of each model, including the HighRes-net model. This study serves as a foundation for further extensive research into the applications of AI in fetal brain fMRI segmentation.
nnDetection for Intracranial Aneurysms Detection and Localization
May 22, 2023Abstract:Intracranial aneurysms are a commonly occurring and life-threatening condition, affecting approximately 3.2% of the general population. Consequently, detecting these aneurysms plays a crucial role in their management. Lesion detection involves the simultaneous localization and categorization of abnormalities within medical images. In this study, we employed the nnDetection framework, a self-configuring framework specifically designed for 3D medical object detection, to detect and localize the 3D coordinates of aneurysms effectively. To capture and extract diverse features associated with aneurysms, we utilized TOF-MRA and structural MRI, both obtained from the ADAM dataset. The performance of our proposed deep learning model was assessed through the utilization of free-response receiver operative characteristics for evaluation purposes. The model's weights and 3D prediction of the bounding box of TOF-MRA are publicly available at https://github.com/orouskhani/AneurysmDetection.
Evolving Tsukamoto Neuro Fuzzy Model for Multiclass Covid 19 Classification with Chest X Ray Images
May 17, 2023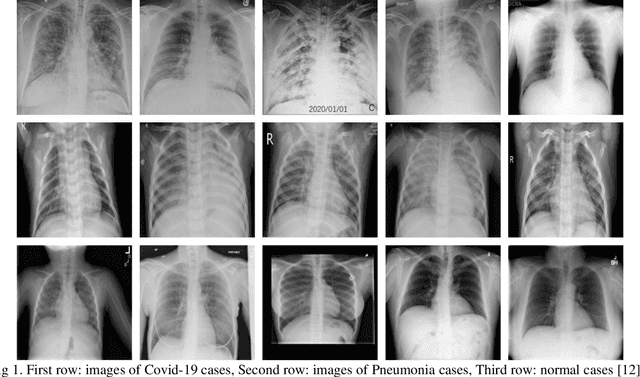
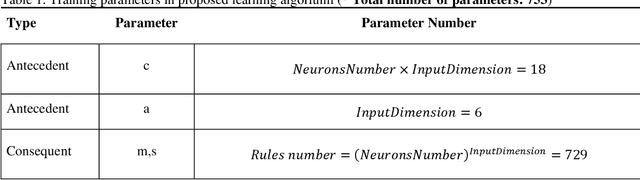
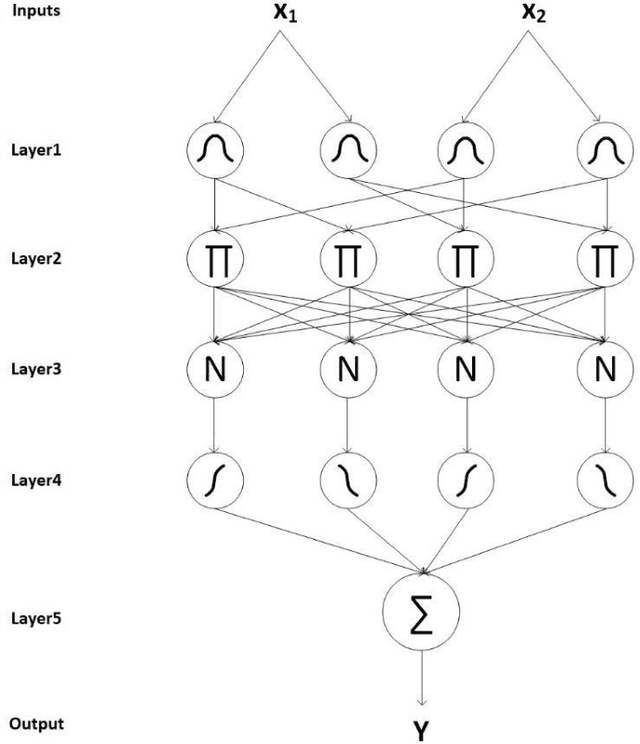
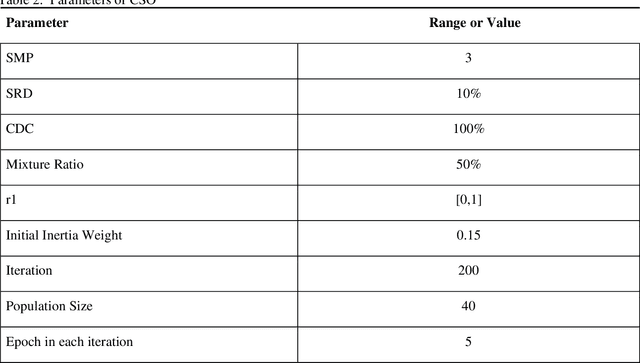
Abstract:Du e to rapid population growth and the need to use artificial intelligence to make quick decisions, developing a machine learning-based disease detection model and abnormality identification system has greatly improved the level of medical diagnosis Since COVID-19 has become one of the most severe diseases in the world, developing an automatic COVID-19 detection framework helps medical doctors in the diagnostic process of disease and provides correct and fast results. In this paper, we propose a machine lear ning based framework for the detection of Covid 19. The proposed model employs a Tsukamoto Neuro Fuzzy Inference network to identify and distinguish Covid 19 disease from normal and pneumonia cases. While the traditional training methods tune the parameters of the neuro-fuzzy model by gradient-based algorithms and recursive least square method, we use an evolutionary-based optimization, the Cat swarm algorithm to update the parameters. In addition, six texture features extracted from chest X-ray images are give n as input to the model. Finally, the proposed model is conducted on the chest X-ray dataset to detect Covid 19. The simulation results indicate that the proposed model achieves an accuracy of 98.51%, sensitivity of 98.35%, specificity of 98.08%, and F1 score of 98.17%.
Generative Adversarial Networks for Brain Images Synthesis: A Review
May 16, 2023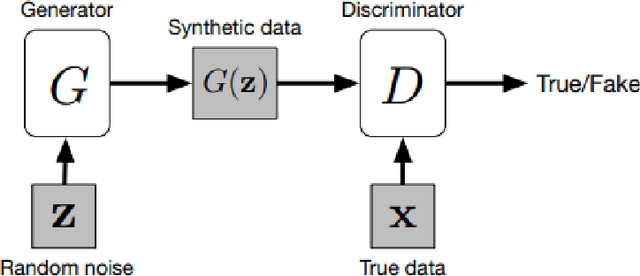
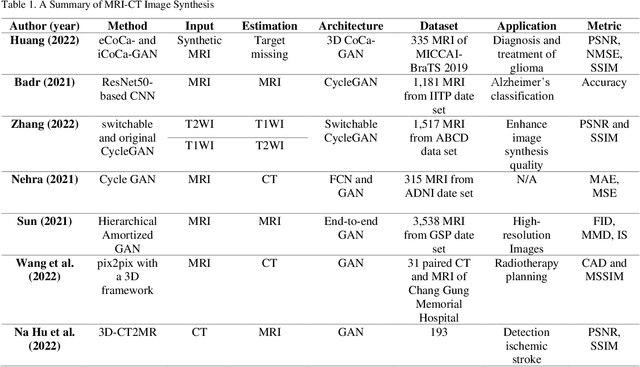
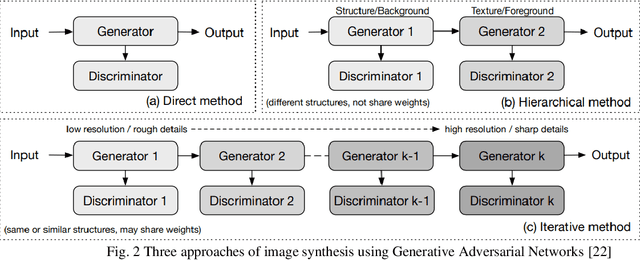
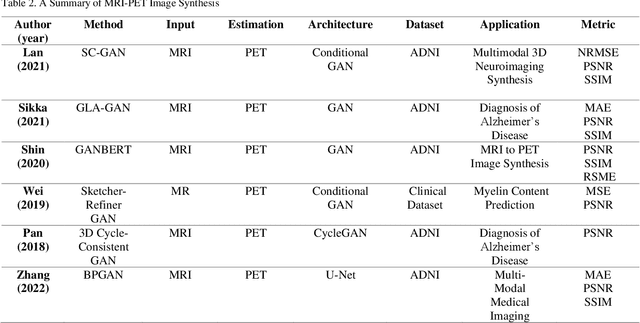
Abstract:In medical imaging, image synthesis is the estimation process of one image (sequence, modality) from another image (sequence, modality). Since images with different modalities provide diverse biomarkers and capture various features, multi-modality imaging is crucial in medicine. While multi-screening is expensive, costly, and time-consuming to report by radiologists, image synthesis methods are capable of artificially generating missing modalities. Deep learning models can automatically capture and extract the high dimensional features. Especially, generative adversarial network (GAN) as one of the most popular generative-based deep learning methods, uses convolutional networks as generators, and estimated images are discriminated as true or false based on a discriminator network. This review provides brain image synthesis via GANs. We summarized the recent developments of GANs for cross-modality brain image synthesis including CT to PET, CT to MRI, MRI to PET, and vice versa.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge