Matthis Synofzik
Identifying latent disease factors differently expressed in patient subgroups using group factor analysis
Oct 10, 2024



Abstract:In this study, we propose a novel approach to uncover subgroup-specific and subgroup-common latent factors addressing the challenges posed by the heterogeneity of neurological and mental disorders, which hinder disease understanding, treatment development, and outcome prediction. The proposed approach, sparse Group Factor Analysis (GFA) with regularised horseshoe priors, was implemented with probabilistic programming and can uncover associations (or latent factors) among multiple data modalities differentially expressed in sample subgroups. Synthetic data experiments showed the robustness of our sparse GFA by correctly inferring latent factors and model parameters. When applied to the Genetic Frontotemporal Dementia Initiative (GENFI) dataset, which comprises patients with frontotemporal dementia (FTD) with genetically defined subgroups, the sparse GFA identified latent disease factors differentially expressed across the subgroups, distinguishing between "subgroup-specific" latent factors within homogeneous groups and "subgroup common" latent factors shared across subgroups. The latent disease factors captured associations between brain structure and non-imaging variables (i.e., questionnaires assessing behaviour and disease severity) across the different genetic subgroups, offering insights into disease profiles. Importantly, two latent factors were more pronounced in the two more homogeneous FTD patient subgroups (progranulin (GRN) and microtubule-associated protein tau (MAPT) mutation), showcasing the method's ability to reveal subgroup-specific characteristics. These findings underscore the potential of sparse GFA for integrating multiple data modalities and identifying interpretable latent disease factors that can improve the characterization and stratification of patients with neurological and mental health disorders.
Early Recognition of Ball Catching Success in Clinical Trials with RNN-Based Predictive Classification
Jul 06, 2021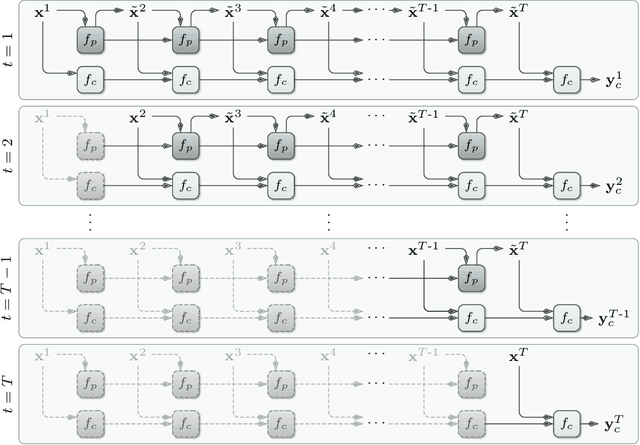
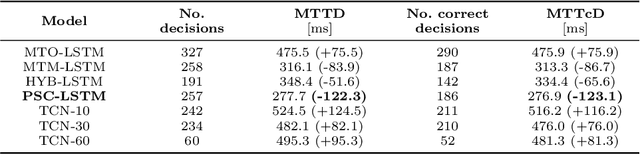
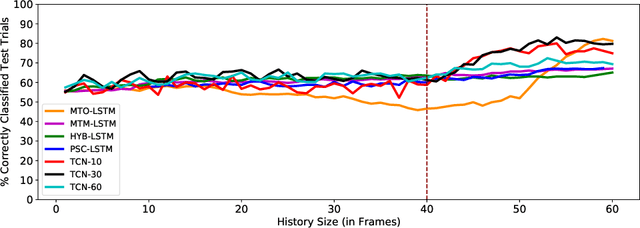

Abstract:Motor disturbances can affect the interaction with dynamic objects, such as catching a ball. A classification of clinical catching trials might give insight into the existence of pathological alterations in the relation of arm and ball movements. Accurate, but also early decisions are required to classify a catching attempt before the catcher's first ball contact. To obtain clinically valuable results, a significant decision confidence of at least 75% is required. Hence, three competing objectives have to be optimized at the same time: accuracy, earliness and decision-making confidence. Here we propose a coupled classification and prediction approach for early time series classification: a predictive, generative recurrent neural network (RNN) forecasts the next data points of ball trajectories based on already available observations; a discriminative RNN continuously generates classification guesses based on the available data points and the unrolled sequence predictions. We compare our approach, which we refer to as predictive sequential classification (PSC), to state-of-the-art sequence learners, including various RNN and temporal convolutional network (TCN) architectures. On this hard real-world task we can consistently demonstrate the superiority of PSC over all other models in terms of accuracy and confidence with respect to earliness of recognition. Specifically, PSC is able to confidently classify the success of catching trials as early as 123 milliseconds before the first ball contact. We conclude that PSC is a promising approach for early time series classification, when accurate and confident decisions are required.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge