Mark Chiew
Low-Rank Conjugate Gradient-Net for Accelerated Cardiac MR Imaging
Nov 17, 2024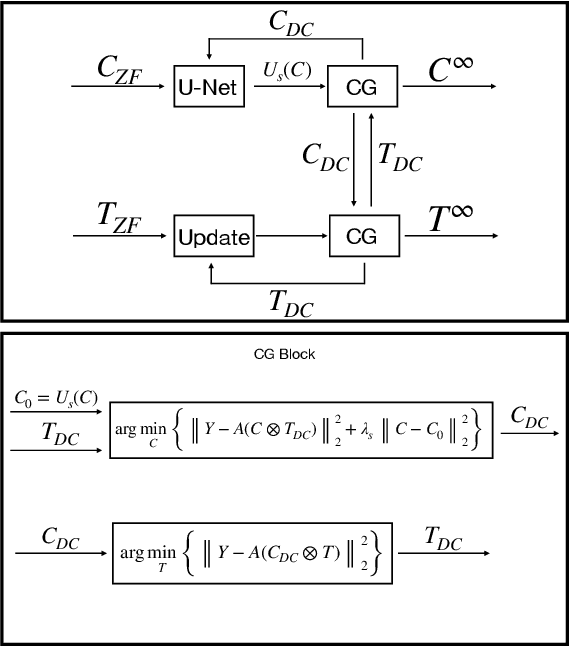
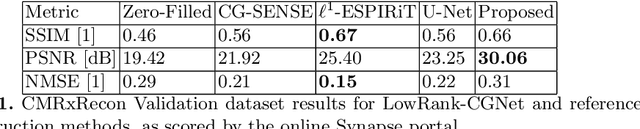

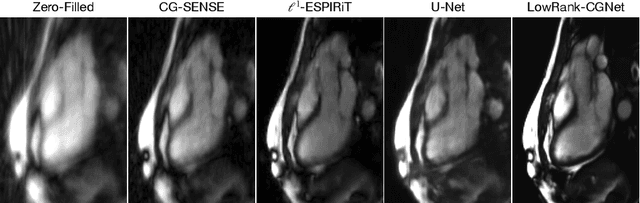
Abstract:Cardiovascular diseases (CVDs) remain the leading cause of mortality and morbidity worldwide. Both diagnosis and prognosis of these diseases benefit from high-quality imaging, which cardiac magnetic resonance imaging provides. CMR imaging requires lengthy acquisition times and multiple breath-holds for a complete exam, which can lead to patient discomfort and frequently results in image artifacts. In this work, we present a Low-rank tensor U-Net method (LowRank-CGNet) that rapidly reconstructs highly undersampled data with a variety of anatomy, contrast, and undersampling artifacts. The model uses conjugate gradient data consistency to solve for the spatial and temporal bases and employs a U-Net to further regularize the basis vectors. Currently, model performance is superior to a standard U-Net, but inferior to conventional compressed sensing methods. In the future, we aim to further improve model performance by increasing the U-Net size, extending the training duration, and dynamically updating the tensor rank for different anatomies.
Self-navigated 3D diffusion MRI using an optimized CAIPI sampling and structured low-rank reconstruction
Jan 11, 2024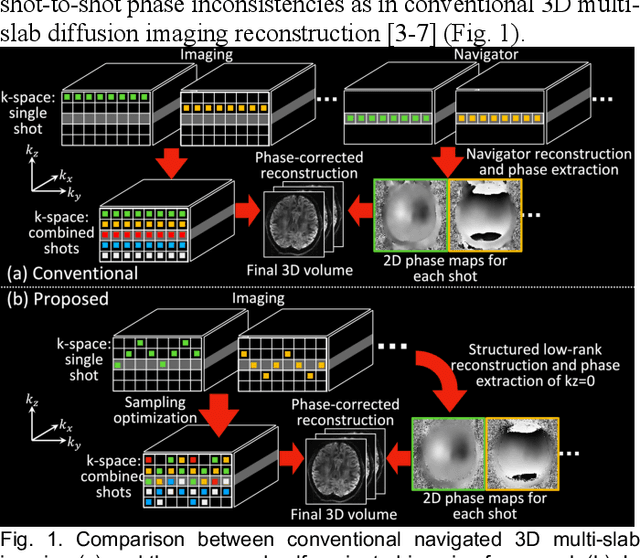
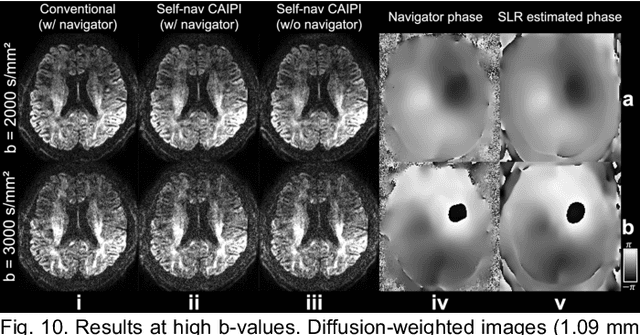
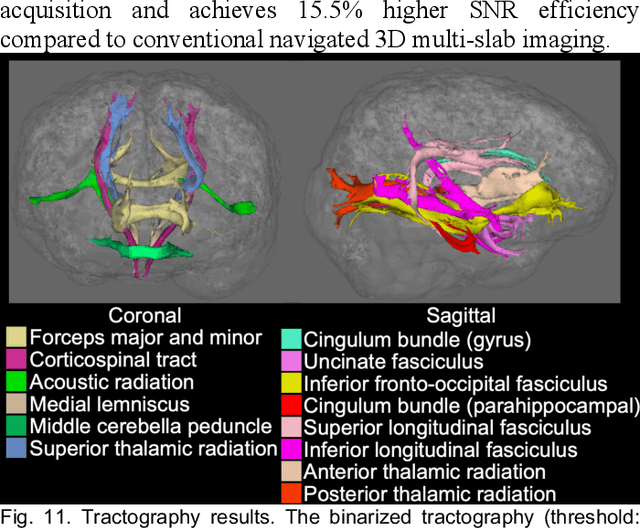
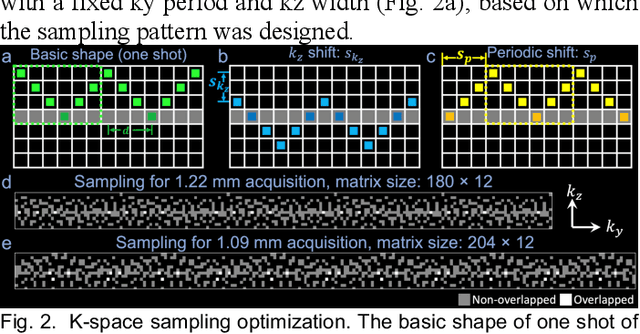
Abstract:3D multi-slab acquisitions are an appealing approach for diffusion MRI because they are compatible with the imaging regime delivering optimal SNR efficiency. In conventional 3D multi-slab imaging, shot-to-shot phase variations caused by motion pose challenges due to the use of multi-shot k-space acquisition. Navigator acquisition after each imaging echo is typically employed to correct phase variations, which prolongs scan time and increases the specific absorption rate (SAR). The aim of this study is to develop a highly efficient, self-navigated method to correct for phase variations in 3D multi-slab diffusion MRI without explicitly acquiring navigators. The sampling of each shot is carefully designed to intersect with the central kz plane of each slab, and the multi-shot sampling is optimized for self-navigation performance while retaining decent reconstruction quality. The central kz intersections from all shots are jointly used to reconstruct a 2D phase map for each shot using a structured low-rank constrained reconstruction that leverages the redundancy in shot and coil dimensions. The phase maps are used to eliminate the shot-to-shot phase inconsistency in the final 3D multi-shot reconstruction. We demonstrate the method's efficacy using retrospective simulations and prospectively acquired in-vivo experiments at 1.22 mm and 1.09 mm isotropic resolutions. Compared to conventional navigated 3D multi-slab imaging, the proposed self-navigated method achieves comparable image quality while shortening the scan time by 31.7% and improving the SNR efficiency by 15.5%. The proposed method produces comparable quality of DTI and white matter tractography to conventional navigated 3D multi-slab acquisition with a much shorter scan time.
Simultaneous self-supervised reconstruction and denoising of sub-sampled MRI data with Noisier2Noise
Oct 07, 2022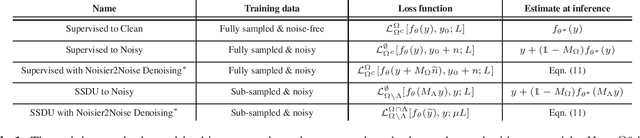



Abstract:Most existing methods for Magnetic Resonance Imaging (MRI) reconstruction with deep learning assume that a high signal-to-noise ratio (SNR), fully sampled sampled dataset exists and use fully supervised training. In many circumstances, however, such a dataset does not exist and may be highly impractical to acquire. Recently, a number of self-supervised methods for MR reconstruction have been proposed, which require a training dataset with sub-sampled k-space data only. However, existing methods do not denoise sampled data, so are only applicable in the high SNR regime. In this work, we propose a method based on Noisier2Noise and Self-Supervised Learning via Data Undersampling (SSDU) that trains a network to reconstruct clean images from sub-sampled, noisy training data. To our knowledge, our approach is the first that simultaneously denoises and reconstructs images in an entirely self-supervised manner. Our method is applicable to any network architecture, has a strong mathematical basis, and is straight-forward to implement. We evaluate our method on the multi-coil fastMRI brain dataset and find that it performs competitively with a network trained on clean, fully sampled data and substantially improves over methods that do not remove measurement noise.
Self-supervised deep learning MRI reconstruction with Noisier2Noise
May 20, 2022
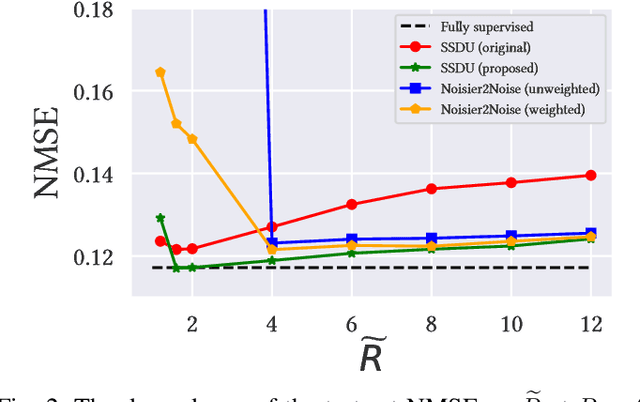

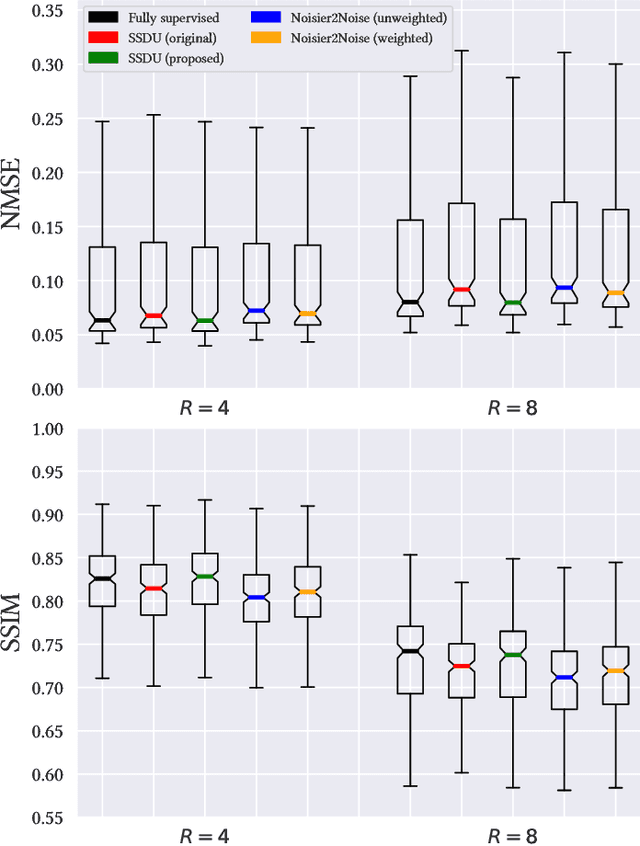
Abstract:In recent years, there has been attention on leveraging the statistical modeling capabilities of neural networks for reconstructing sub-sampled Magnetic Resonance Imaging (MRI) data. Most proposed methods assume the existence of a representative fully-sampled dataset and use fully-supervised training. However, for many applications, fully sampled training data is not available, and may be highly impractical to acquire. The development of self-supervised methods, which use only sub-sampled data for training, are therefore highly desirable. This work extends the Noisier2Noise framework, which was originally constructed for self-supervised denoising tasks, to variable density sub-sampled MRI data. Further, we use the Noisier2Noise framework to analytically explain the performance of Self-Supervised Learning via Data Undersampling (SSDU), a recently proposed method that performs well in practice but until now lacked theoretical justification. We also use Noisier2Noise to propose a modification of SSDU that we find substantially improves its reconstruction quality and robustness, offering a test set mean-squared-error within 1% of fully supervised training on the fastMRI brain dataset.
Tuning-free multi-coil compressed sensing MRI with Parallel Variable Density Approximate Message Passing
Mar 08, 2022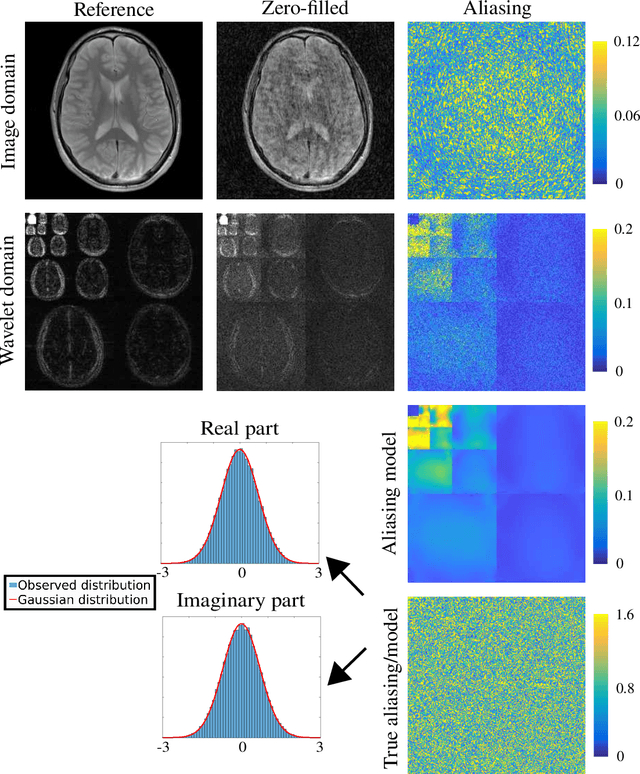
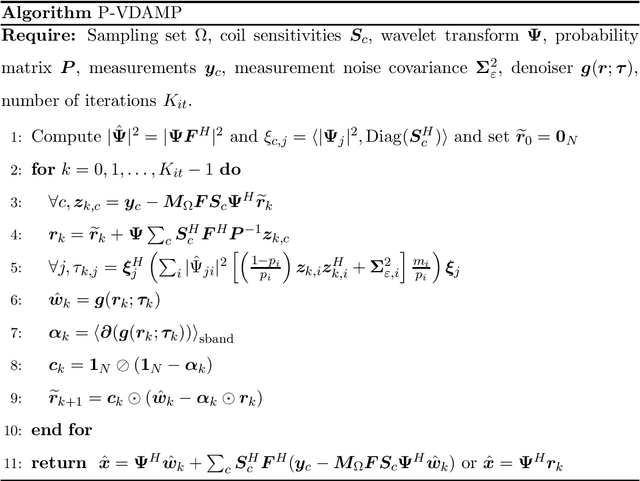
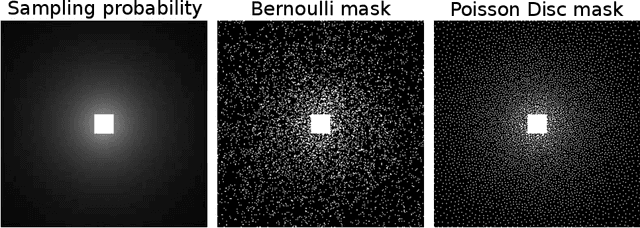
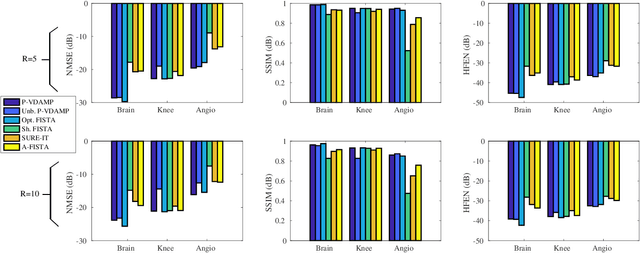
Abstract:Purpose: To develop a tuning-free method for multi-coil compressed sensing MRI that performs competitively with algorithms with an optimally tuned sparse parameter. Theory: The Parallel Variable Density Approximate Message Passing (P-VDAMP) algorithm is proposed. For Bernoulli random variable density sampling, P-VDAMP obeys a "state evolution", where the intermediate per-iteration image estimate is distributed according to the ground truth corrupted by a Gaussian vector with approximately known covariance. State evolution is leveraged to automatically tune sparse parameters on-the-fly with Stein's Unbiased Risk Estimate (SURE). Methods: P-VDAMP is evaluated on brain, knee and angiogram datasets at acceleration factors 5 and 10 and compared with four variants of the Fast Iterative Shrinkage-Thresholding algorithm (FISTA), including two tuning-free variants from the literature. Results: The proposed method is found to have a similar reconstruction quality and time to convergence as FISTA with an optimally tuned sparse weighting. Conclusions: P-VDAMP is an efficient, robust and principled method for on-the-fly parameter tuning that is competitive with optimally tuned FISTA and offers substantial robustness and reconstruction quality improvements over competing tuning-free methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge