Charles Millard
Simultaneous self-supervised reconstruction and denoising of sub-sampled MRI data with Noisier2Noise
Oct 07, 2022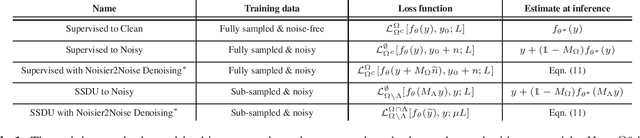



Abstract:Most existing methods for Magnetic Resonance Imaging (MRI) reconstruction with deep learning assume that a high signal-to-noise ratio (SNR), fully sampled sampled dataset exists and use fully supervised training. In many circumstances, however, such a dataset does not exist and may be highly impractical to acquire. Recently, a number of self-supervised methods for MR reconstruction have been proposed, which require a training dataset with sub-sampled k-space data only. However, existing methods do not denoise sampled data, so are only applicable in the high SNR regime. In this work, we propose a method based on Noisier2Noise and Self-Supervised Learning via Data Undersampling (SSDU) that trains a network to reconstruct clean images from sub-sampled, noisy training data. To our knowledge, our approach is the first that simultaneously denoises and reconstructs images in an entirely self-supervised manner. Our method is applicable to any network architecture, has a strong mathematical basis, and is straight-forward to implement. We evaluate our method on the multi-coil fastMRI brain dataset and find that it performs competitively with a network trained on clean, fully sampled data and substantially improves over methods that do not remove measurement noise.
Self-supervised deep learning MRI reconstruction with Noisier2Noise
May 20, 2022
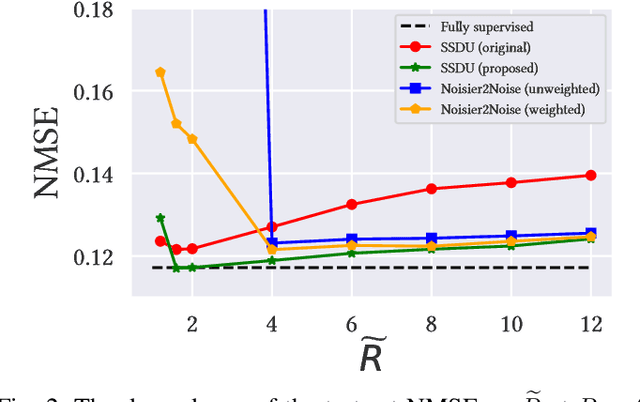

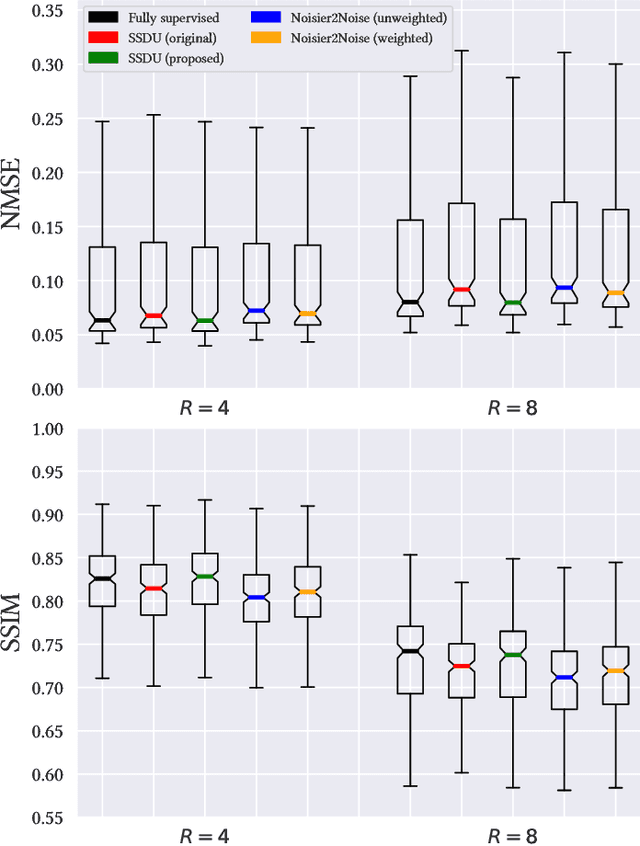
Abstract:In recent years, there has been attention on leveraging the statistical modeling capabilities of neural networks for reconstructing sub-sampled Magnetic Resonance Imaging (MRI) data. Most proposed methods assume the existence of a representative fully-sampled dataset and use fully-supervised training. However, for many applications, fully sampled training data is not available, and may be highly impractical to acquire. The development of self-supervised methods, which use only sub-sampled data for training, are therefore highly desirable. This work extends the Noisier2Noise framework, which was originally constructed for self-supervised denoising tasks, to variable density sub-sampled MRI data. Further, we use the Noisier2Noise framework to analytically explain the performance of Self-Supervised Learning via Data Undersampling (SSDU), a recently proposed method that performs well in practice but until now lacked theoretical justification. We also use Noisier2Noise to propose a modification of SSDU that we find substantially improves its reconstruction quality and robustness, offering a test set mean-squared-error within 1% of fully supervised training on the fastMRI brain dataset.
Tuning-free multi-coil compressed sensing MRI with Parallel Variable Density Approximate Message Passing
Mar 08, 2022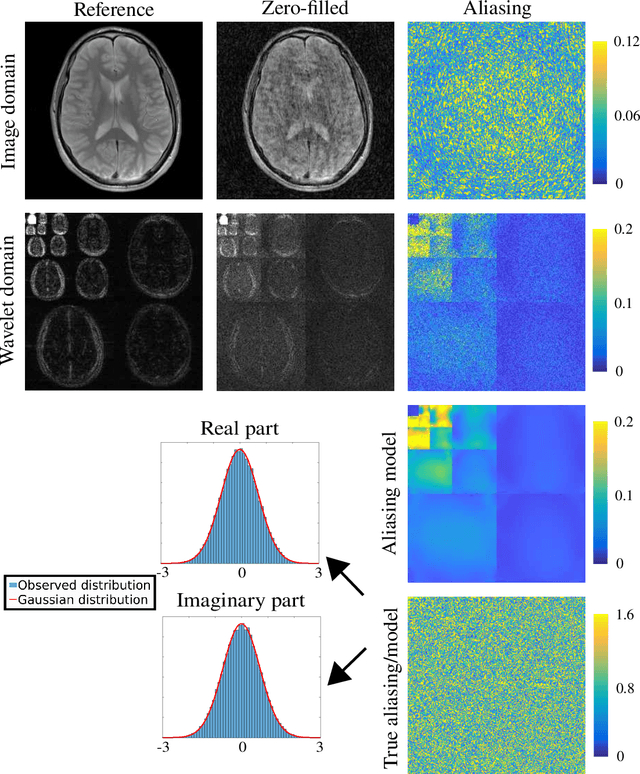
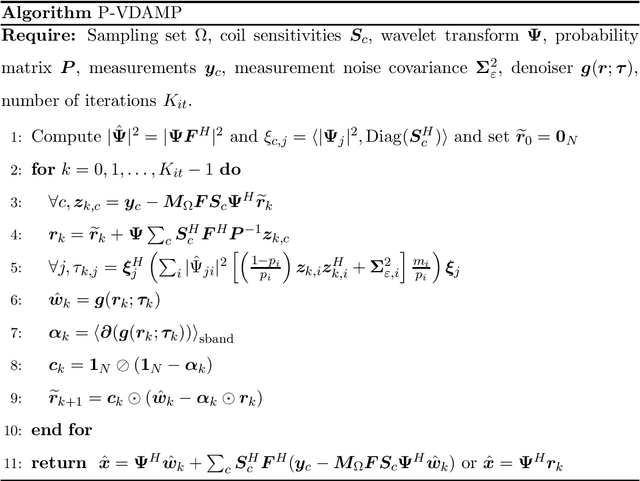
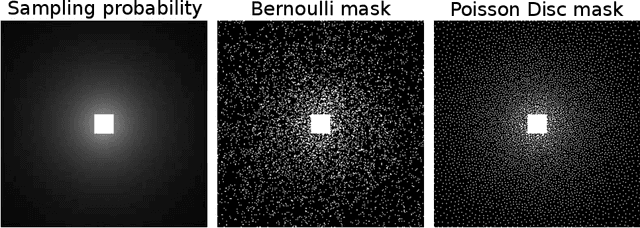
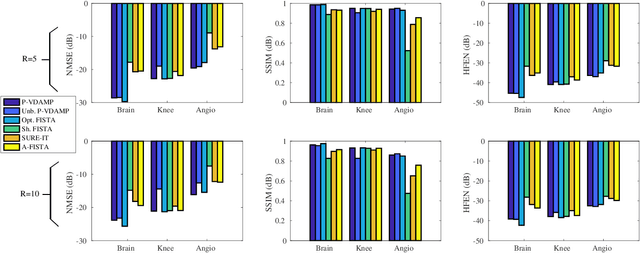
Abstract:Purpose: To develop a tuning-free method for multi-coil compressed sensing MRI that performs competitively with algorithms with an optimally tuned sparse parameter. Theory: The Parallel Variable Density Approximate Message Passing (P-VDAMP) algorithm is proposed. For Bernoulli random variable density sampling, P-VDAMP obeys a "state evolution", where the intermediate per-iteration image estimate is distributed according to the ground truth corrupted by a Gaussian vector with approximately known covariance. State evolution is leveraged to automatically tune sparse parameters on-the-fly with Stein's Unbiased Risk Estimate (SURE). Methods: P-VDAMP is evaluated on brain, knee and angiogram datasets at acceleration factors 5 and 10 and compared with four variants of the Fast Iterative Shrinkage-Thresholding algorithm (FISTA), including two tuning-free variants from the literature. Results: The proposed method is found to have a similar reconstruction quality and time to convergence as FISTA with an optimally tuned sparse weighting. Conclusions: P-VDAMP is an efficient, robust and principled method for on-the-fly parameter tuning that is competitive with optimally tuned FISTA and offers substantial robustness and reconstruction quality improvements over competing tuning-free methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge