Manuel Morante
Latent Mode Decomposition
May 23, 2025Abstract:We introduce Variational Latent Mode Decomposition (VLMD), a new algorithm for extracting oscillatory modes and associated connectivity structures from multivariate signals. VLMD addresses key limitations of existing Multivariate Mode Decomposition (MMD) techniques -including high computational cost, sensitivity to parameter choices, and weak modeling of interchannel dependencies. Its improved performance is driven by a novel underlying model, Latent Mode Decomposition (LMD), which blends sparse coding and mode decomposition to represent multichannel signals as sparse linear combinations of shared latent components composed of AM-FM oscillatory modes. This formulation enables VLMD to operate in a lower-dimensional latent space, enhancing robustness to noise, scalability, and interpretability. The algorithm solves a constrained variational optimization problem that jointly enforces reconstruction fidelity, sparsity, and frequency regularization. Experiments on synthetic and real-world datasets demonstrate that VLMD outperforms state-of-the-art MMD methods in accuracy, efficiency, and interpretability of extracted structures.
A probabilistic estimation of remaining useful life from censored time-to-event data
May 02, 2024Abstract:Predicting the remaining useful life (RUL) of ball bearings plays an important role in predictive maintenance. A common definition of the RUL is the time until a bearing is no longer functional, which we denote as an event, and many data-driven methods have been proposed to predict the RUL. However, few studies have addressed the problem of censored data, where this event of interest is not observed, and simply ignoring these observations can lead to an overestimation of the failure risk. In this paper, we propose a probabilistic estimation of RUL using survival analysis that supports censored data. First, we analyze sensor readings from ball bearings in the frequency domain and annotate when a bearing starts to deteriorate by calculating the Kullback-Leibler (KL) divergence between the probability density function (PDF) of the current process and a reference PDF. Second, we train several survival models on the annotated bearing dataset, capable of predicting the RUL over a finite time horizon using the survival function. This function is guaranteed to be strictly monotonically decreasing and is an intuitive estimation of the remaining lifetime. We demonstrate our approach in the XJTU-SY dataset using cross-validation and find that Random Survival Forests consistently outperforms both non-neural networks and neural networks in terms of the mean absolute error (MAE). Our work encourages the inclusion of censored data in predictive maintenance models and highlights the unique advantages that survival analysis offers when it comes to probabilistic RUL estimation and early fault detection.
A lite parametric model for the Hemodynamic Response Function
Apr 28, 2020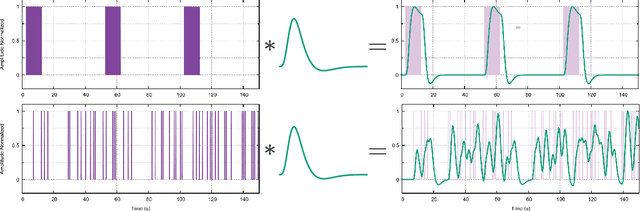
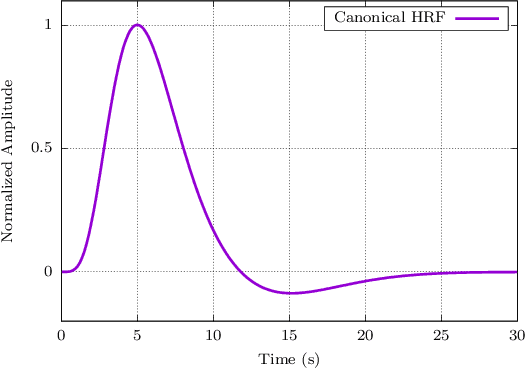
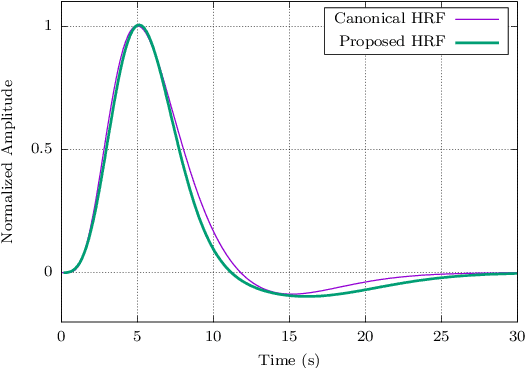
Abstract:When working with task-related fMRI data, one of the most crucial parts of the data analysis consists of determining a proper estimate of the BOLD response. The following document presents a lite model for the Hemodynamic Response Function HRF. Between other advances, the proposed model present less number of parameters compared to other similar HRF alternative, which reduces its optimization complexity and facilitates its potential applications.
Information Assisted Dictionary Learning for fMRI data analysis
May 11, 2018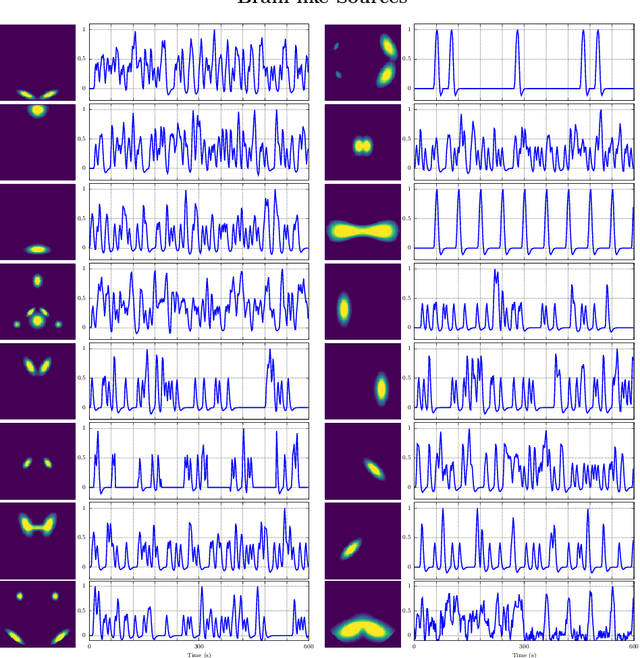
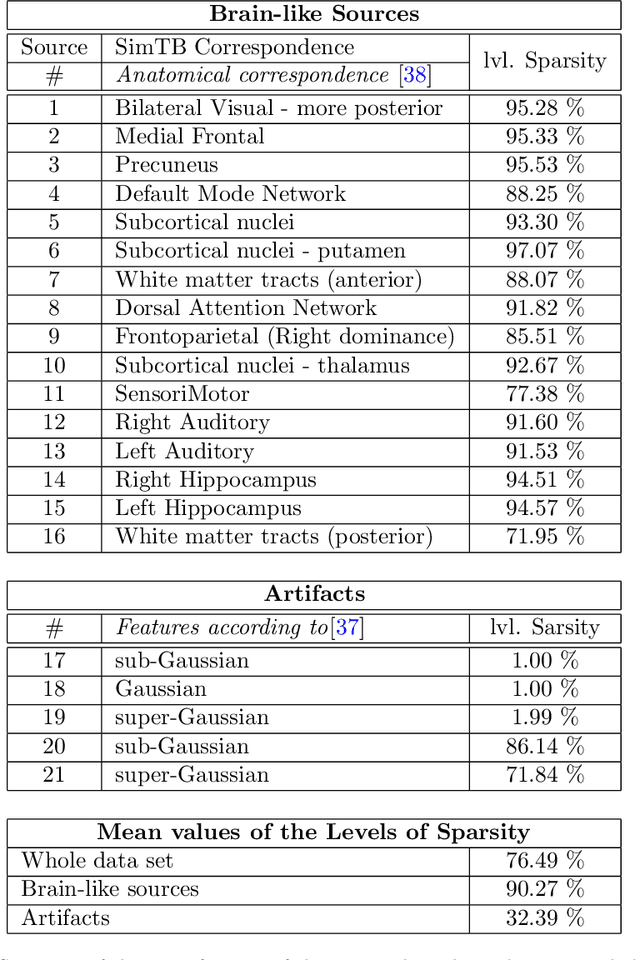
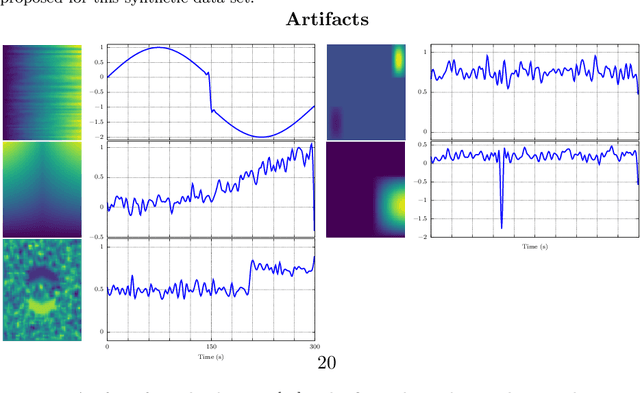

Abstract:In this paper, the task-related fMRI problem is treated in its matrix factorization formulation. The focus of the reported work is on the dictionary learning (DL) matrix factorization approach. A major novelty of the paper lies in the incorporation of well-established assumptions associated with the GLM technique, which is currently in use by the neuroscientists. These assumptions are embedded as constraints in the DL formulation. In this way, our approach provides a framework of combining well-established and understood techniques with a more ``modern'' and powerful tool. Furthermore, this paper offers a way to relax a major drawback associated with DL techniques; that is, the proper tuning of the DL regularization parameter. This parameter plays a critical role in DL-based fMRI analysis since it essentially determines the shape and structures of the estimated functional brain networks. However, in actual fMRI data analysis, the lack of ground truth renders the a priori choice of the regularization parameter a truly challenging task. Indeed, the values of the DL regularization parameter, associated with the $\ell_1$ sparsity promoting norm, do not convey any tangible physical meaning. So it is practically difficult to guess its proper value. In this paper, the DL problem is reformulated around a sparsity-promoting constraint that can directly be related to the minimum amount of voxels that the spatial maps of the functional brain networks occupy. Such information is documented and it is readily available to neuroscientists and experts in the field. The proposed method is tested against a number of other popular techniques and the obtained performance gains are reported using a number of synthetic fMRI data. Results with real data have also been obtained in the context of a number of experiments and will be soon reported in a different publication.
Unsupervised learning of the brain connectivity dynamic using residual D-net
Apr 20, 2018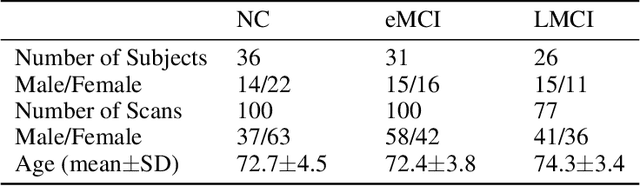
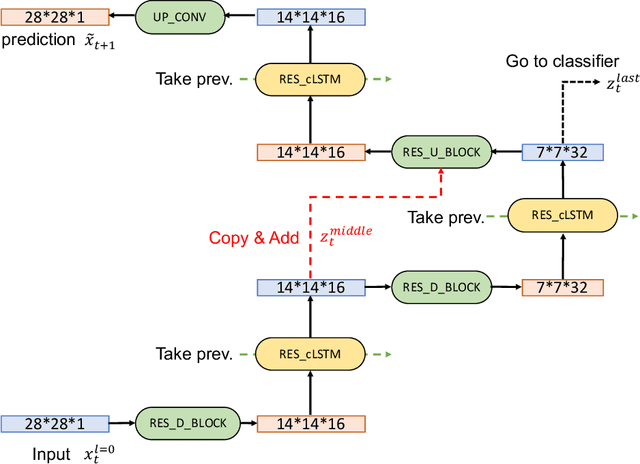
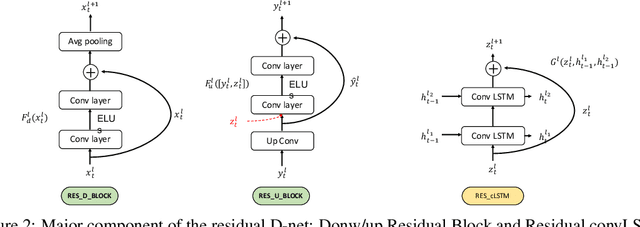
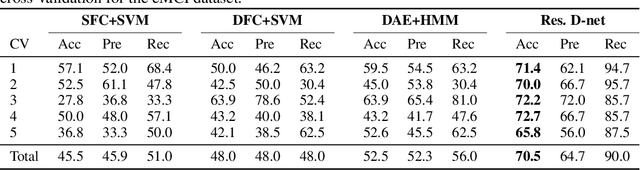
Abstract:In this paper, we propose a novel unsupervised learning method to learn the brain dynamics using a deep learning architecture named residual D-net. As it is often the case in medical research, in contrast to typical deep learning tasks, the size of the resting-state functional Magnetic Resonance Image (rs-fMRI) datasets for training is limited. Thus, the available data should be very efficiently used to learn the complex patterns underneath the brain connectivity dynamics. To address this issue, we use residual connections to alleviate the training complexity through recurrent multi-scale representation. We conduct two classification tasks to differentiate early and late stage Mild Cognitive Impairment (MCI) from Normal healthy Control (NC) subjects. The experiments verify that our proposed residual D-net indeed learns the brain connectivity dynamics, leading to significantly higher classification accuracy compared to previously published techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge