Mackenzie Weygandt Mathis
FMPose3D: monocular 3D pose estimation via flow matching
Feb 05, 2026Abstract:Monocular 3D pose estimation is fundamentally ill-posed due to depth ambiguity and occlusions, thereby motivating probabilistic methods that generate multiple plausible 3D pose hypotheses. In particular, diffusion-based models have recently demonstrated strong performance, but their iterative denoising process typically requires many timesteps for each prediction, making inference computationally expensive. In contrast, we leverage Flow Matching (FM) to learn a velocity field defined by an Ordinary Differential Equation (ODE), enabling efficient generation of 3D pose samples with only a few integration steps. We propose a novel generative pose estimation framework, FMPose3D, that formulates 3D pose estimation as a conditional distribution transport problem. It continuously transports samples from a standard Gaussian prior to the distribution of plausible 3D poses conditioned only on 2D inputs. Although ODE trajectories are deterministic, FMPose3D naturally generates various pose hypotheses by sampling different noise seeds. To obtain a single accurate prediction from those hypotheses, we further introduce a Reprojection-based Posterior Expectation Aggregation (RPEA) module, which approximates the Bayesian posterior expectation over 3D hypotheses. FMPose3D surpasses existing methods on the widely used human pose estimation benchmarks Human3.6M and MPI-INF-3DHP, and further achieves state-of-the-art performance on the 3D animal pose datasets Animal3D and CtrlAni3D, demonstrating strong performance across both 3D pose domains. The code is available at https://github.com/AdaptiveMotorControlLab/FMPose3D.
Time-series attribution maps with regularized contrastive learning
Feb 17, 2025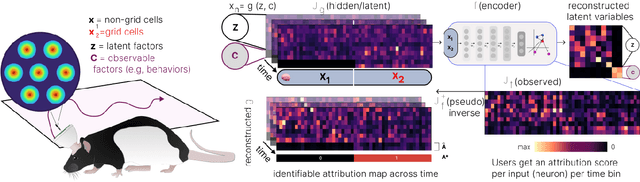
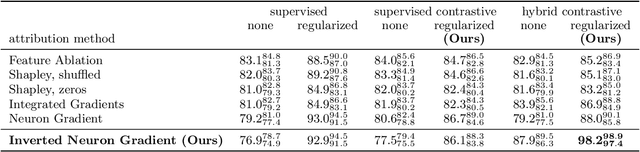
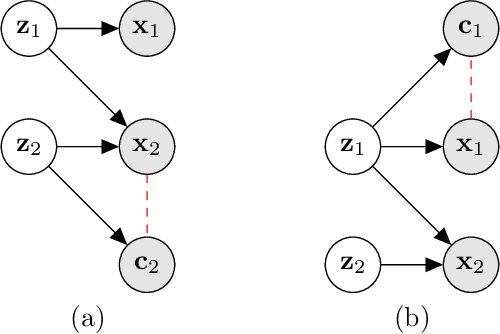
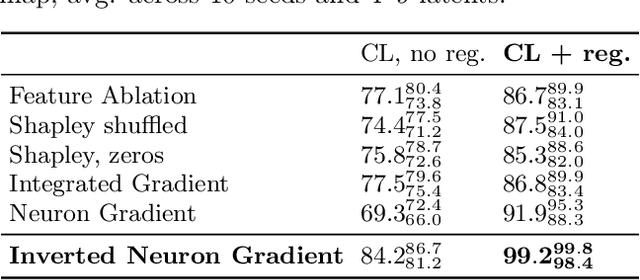
Abstract:Gradient-based attribution methods aim to explain decisions of deep learning models but so far lack identifiability guarantees. Here, we propose a method to generate attribution maps with identifiability guarantees by developing a regularized contrastive learning algorithm trained on time-series data plus a new attribution method called Inverted Neuron Gradient (collectively named xCEBRA). We show theoretically that xCEBRA has favorable properties for identifying the Jacobian matrix of the data generating process. Empirically, we demonstrate robust approximation of zero vs. non-zero entries in the ground-truth attribution map on synthetic datasets, and significant improvements across previous attribution methods based on feature ablation, Shapley values, and other gradient-based methods. Our work constitutes a first example of identifiable inference of time-series attribution maps and opens avenues to a better understanding of time-series data, such as for neural dynamics and decision-processes within neural networks.
* Accepted at The 28th International Conference on Artificial Intelligence and Statistics (AISTATS 2025). Code is available at https://github.com/AdaptiveMotorControlLab/CEBRA
Adaptive Intelligence: leveraging insights from adaptive behavior in animals to build flexible AI systems
Nov 21, 2024



Abstract:Biological intelligence is inherently adaptive -- animals continually adjust their actions based on environmental feedback. However, creating adaptive artificial intelligence (AI) remains a major challenge. The next frontier is to go beyond traditional AI to develop "adaptive intelligence," defined here as harnessing insights from biological intelligence to build agents that can learn online, generalize, and rapidly adapt to changes in their environment. Recent advances in neuroscience offer inspiration through studies that increasingly focus on how animals naturally learn and adapt their world models. In this Perspective, I will review the behavioral and neural foundations of adaptive biological intelligence, the parallel progress in AI, and explore brain-inspired approaches for building more adaptive algorithms.
Learnable latent embeddings for joint behavioral and neural analysis
Apr 01, 2022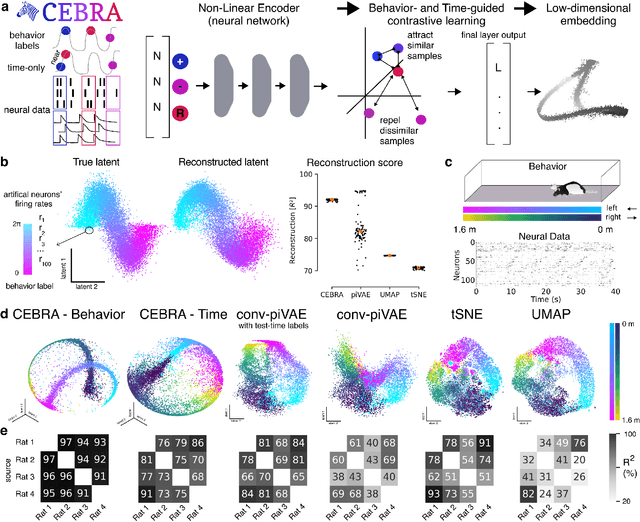
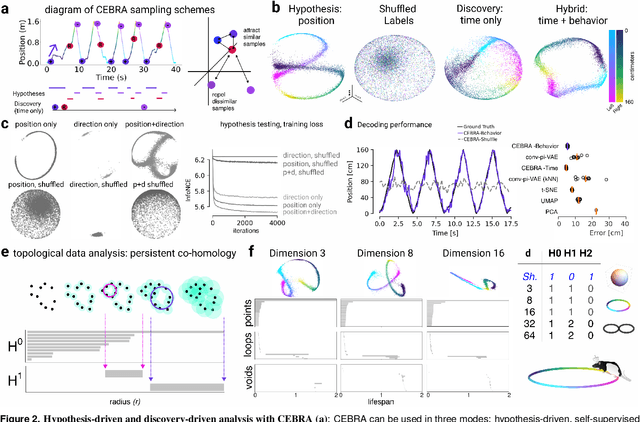
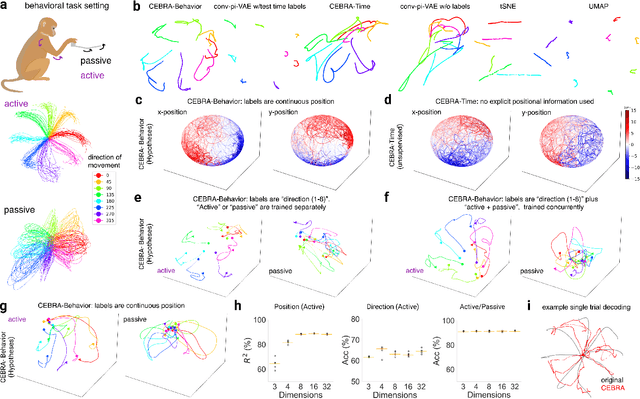
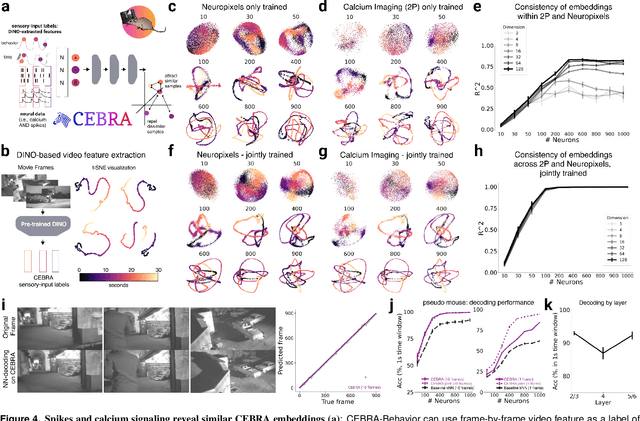
Abstract:Mapping behavioral actions to neural activity is a fundamental goal of neuroscience. As our ability to record large neural and behavioral data increases, there is growing interest in modeling neural dynamics during adaptive behaviors to probe neural representations. In particular, neural latent embeddings can reveal underlying correlates of behavior, yet, we lack non-linear techniques that can explicitly and flexibly leverage joint behavior and neural data. Here, we fill this gap with a novel method, CEBRA, that jointly uses behavioral and neural data in a hypothesis- or discovery-driven manner to produce consistent, high-performance latent spaces. We validate its accuracy and demonstrate our tool's utility for both calcium and electrophysiology datasets, across sensory and motor tasks, and in simple or complex behaviors across species. It allows for single and multi-session datasets to be leveraged for hypothesis testing or can be used label-free. Lastly, we show that CEBRA can be used for the mapping of space, uncovering complex kinematic features, and rapid, high-accuracy decoding of natural movies from visual cortex.
Panoptic animal pose estimators are zero-shot performers
Mar 14, 2022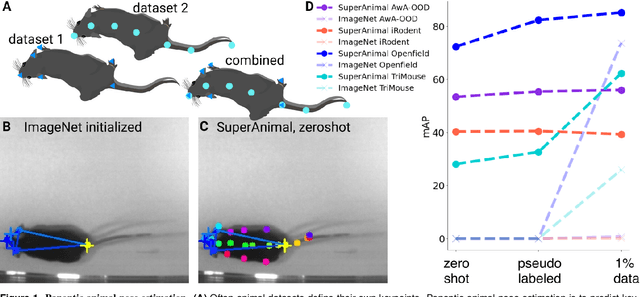
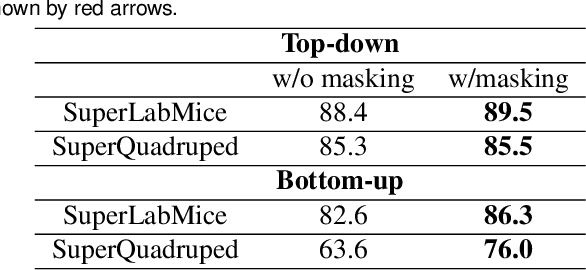
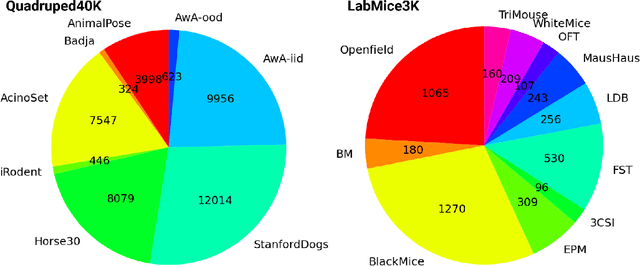
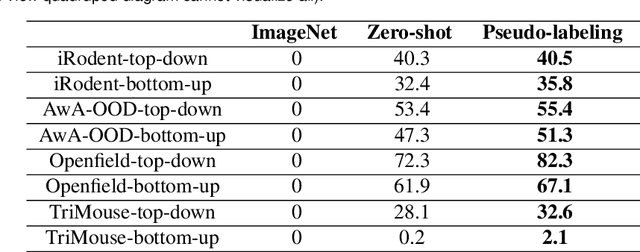
Abstract:Animal pose estimation is critical in applications ranging from life science research, agriculture, to veterinary medicine. Compared to human pose estimation, the performance of animal pose estimation is limited by the size of available datasets and the generalization of a model across datasets. Typically different keypoints are labeled regardless of whether the species are the same or not, leaving animal pose datasets to have disjoint or partially overlapping keypoints. As a consequence, a model cannot be used as a plug-and-play solution across datasets. This reality motivates us to develop panoptic animal pose estimation models that are able to predict keypoints defined in all datasets. In this work we propose a simple yet effective way to merge differentially labeled datasets to obtain the largest quadruped and lab mouse pose dataset. Using a gradient masking technique, so called SuperAnimal-models are able to predict keypoints that are distributed across datasets and exhibit strong zero-shot performance. The models can be further improved by (pseudo) labeled fine-tuning. These models outperform ImageNet-initialized models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge