Markus Frey
Time-series attribution maps with regularized contrastive learning
Feb 17, 2025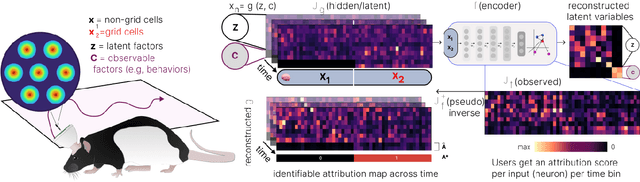
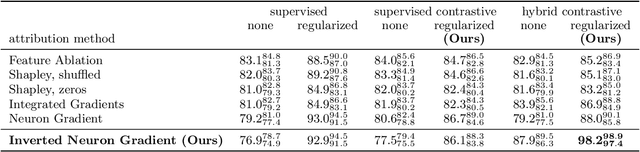
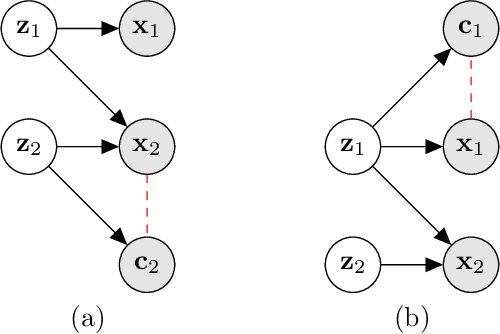
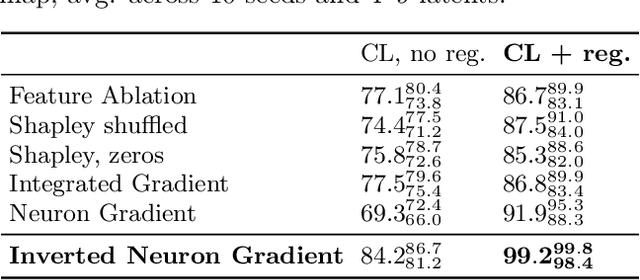
Abstract:Gradient-based attribution methods aim to explain decisions of deep learning models but so far lack identifiability guarantees. Here, we propose a method to generate attribution maps with identifiability guarantees by developing a regularized contrastive learning algorithm trained on time-series data plus a new attribution method called Inverted Neuron Gradient (collectively named xCEBRA). We show theoretically that xCEBRA has favorable properties for identifying the Jacobian matrix of the data generating process. Empirically, we demonstrate robust approximation of zero vs. non-zero entries in the ground-truth attribution map on synthetic datasets, and significant improvements across previous attribution methods based on feature ablation, Shapley values, and other gradient-based methods. Our work constitutes a first example of identifiable inference of time-series attribution maps and opens avenues to a better understanding of time-series data, such as for neural dynamics and decision-processes within neural networks.
* Accepted at The 28th International Conference on Artificial Intelligence and Statistics (AISTATS 2025). Code is available at https://github.com/AdaptiveMotorControlLab/CEBRA
Probing neural representations of scene perception in a hippocampally dependent task using artificial neural networks
Mar 11, 2023



Abstract:Deep artificial neural networks (DNNs) trained through backpropagation provide effective models of the mammalian visual system, accurately capturing the hierarchy of neural responses through primary visual cortex to inferior temporal cortex (IT). However, the ability of these networks to explain representations in higher cortical areas is relatively lacking and considerably less well researched. For example, DNNs have been less successful as a model of the egocentric to allocentric transformation embodied by circuits in retrosplenial and posterior parietal cortex. We describe a novel scene perception benchmark inspired by a hippocampal dependent task, designed to probe the ability of DNNs to transform scenes viewed from different egocentric perspectives. Using a network architecture inspired by the connectivity between temporal lobe structures and the hippocampus, we demonstrate that DNNs trained using a triplet loss can learn this task. Moreover, by enforcing a factorized latent space, we can split information propagation into "what" and "where" pathways, which we use to reconstruct the input. This allows us to beat the state-of-the-art for unsupervised object segmentation on the CATER and MOVi-A,B,C benchmarks.
Memory efficient brain tumor segmentation using an autoencoder-regularized U-Net
Oct 04, 2019



Abstract:Early diagnosis and accurate segmentation of brain tumors are imperative for successful treatment. Unfortunately, manual segmentation is time consuming, costly and despite extensive human expertise often inaccurate. Here, we present an MRI-based tumor segmentation framework using an autoencoder-regularized 3D-convolutional neural network. We trained the model on manually segmented structural T1, T1ce, T2, and Flair MRI images of 335 patients with tumors of variable severity, size and location. We then tested the model using independent data of 125 patients and successfully segmented brain tumors into three subregions: the tumor core (TC), the enhancing tumor (ET) and the whole tumor (WT). We also explored several data augmentations and preprocessing steps to improve segmentation performance. Importantly, our model was implemented on a single NVIDIA GTX1060 graphics unit and hence optimizes tumor segmentation for widely affordable hardware. In sum, we present a memory-efficient and affordable solution to tumor segmentation to support the accurate diagnostics of oncological brain pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge