MacLean P. Nasrallah
BraTS-Path Challenge: Assessing Heterogeneous Histopathologic Brain Tumor Sub-regions
May 17, 2024Abstract:Glioblastoma is the most common primary adult brain tumor, with a grim prognosis - median survival of 12-18 months following treatment, and 4 months otherwise. Glioblastoma is widely infiltrative in the cerebral hemispheres and well-defined by heterogeneous molecular and micro-environmental histopathologic profiles, which pose a major obstacle in treatment. Correctly diagnosing these tumors and assessing their heterogeneity is crucial for choosing the precise treatment and potentially enhancing patient survival rates. In the gold-standard histopathology-based approach to tumor diagnosis, detecting various morpho-pathological features of distinct histology throughout digitized tissue sections is crucial. Such "features" include the presence of cellular tumor, geographic necrosis, pseudopalisading necrosis, areas abundant in microvascular proliferation, infiltration into the cortex, wide extension in subcortical white matter, leptomeningeal infiltration, regions dense with macrophages, and the presence of perivascular or scattered lymphocytes. With these features in mind and building upon the main aim of the BraTS Cluster of Challenges https://www.synapse.org/brats2024, the goal of the BraTS-Path challenge is to provide a systematically prepared comprehensive dataset and a benchmarking environment to develop and fairly compare deep-learning models capable of identifying tumor sub-regions of distinct histologic profile. These models aim to further our understanding of the disease and assist in the diagnosis and grading of conditions in a consistent manner.
Detecting Histologic Glioblastoma Regions of Prognostic Relevance
Feb 01, 2023
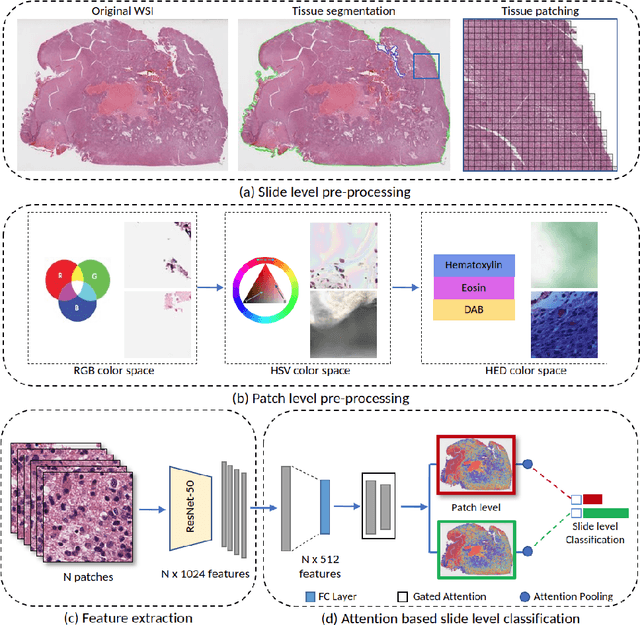
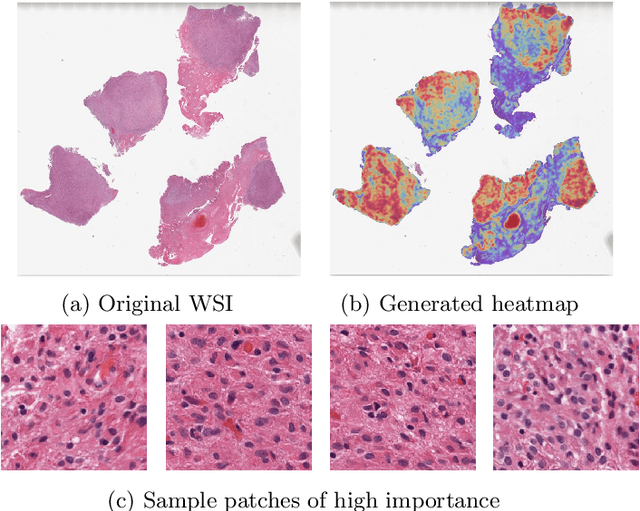
Abstract:Glioblastoma is the most common and aggressive malignant adult tumor of the central nervous system, with grim prognosis and heterogeneous morphologic and molecular profiles. Since the adoption of the current standard of care treatment, 18 years ago, there are no substantial prognostic improvements noticed. Accurate prediction of patient overall survival (OS) from clinical histopathology whole slide images (WSI) using advanced computational methods could contribute to optimization of clinical decision making and patient management. Here, we focus on identifying prognostically relevant glioblastoma morphologic patterns on H&E stained WSI. The exact approach capitalizes on the comprehensive WSI curation of apparent artifactual content and on an interpretability mechanism via a weakly supervised attention based multiple instance learning algorithm that further utilizes clustering to constrain the search space. The automatically identified patterns of high diagnostic value are used to classify the WSI as representative of a short or a long survivor. Identifying tumor morphologic patterns associated with short and long OS will allow the clinical neuropathologist to provide additional prognostic information gleaned during microscopic assessment to the treating team, as well as suggest avenues of biological investigation for understanding and potentially treating glioblastoma.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge