Garv Mehdiratta
Detecting Histologic Glioblastoma Regions of Prognostic Relevance
Feb 01, 2023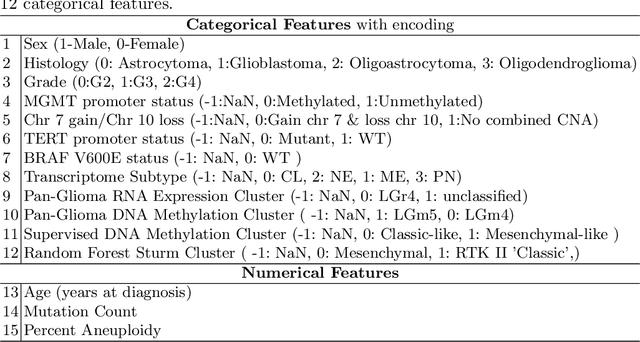
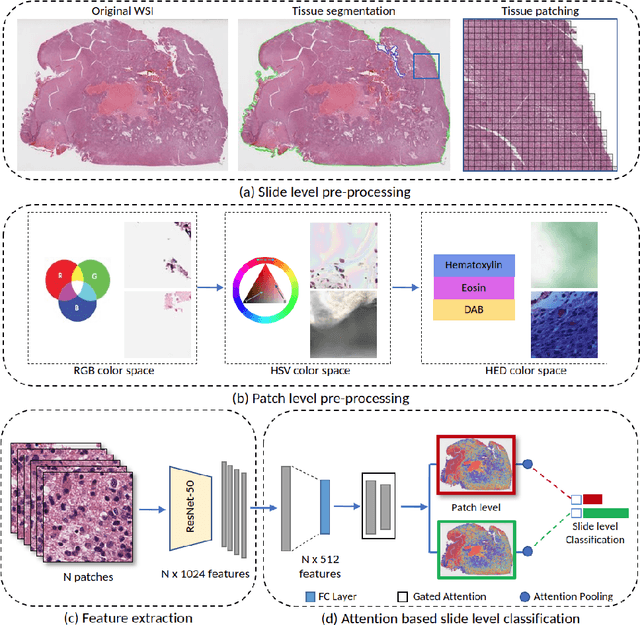
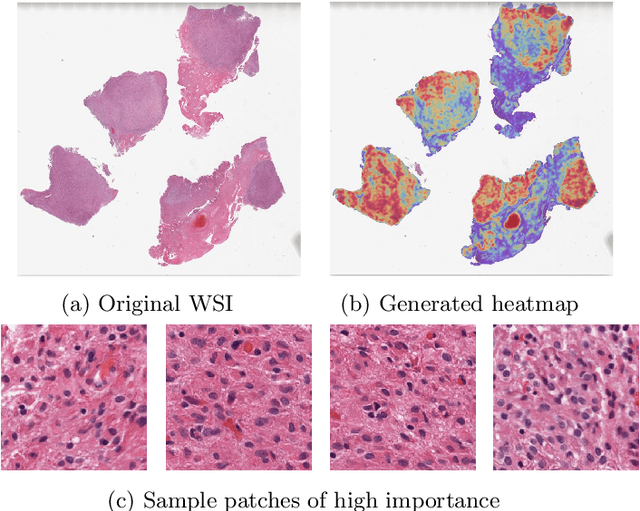
Abstract:Glioblastoma is the most common and aggressive malignant adult tumor of the central nervous system, with grim prognosis and heterogeneous morphologic and molecular profiles. Since the adoption of the current standard of care treatment, 18 years ago, there are no substantial prognostic improvements noticed. Accurate prediction of patient overall survival (OS) from clinical histopathology whole slide images (WSI) using advanced computational methods could contribute to optimization of clinical decision making and patient management. Here, we focus on identifying prognostically relevant glioblastoma morphologic patterns on H&E stained WSI. The exact approach capitalizes on the comprehensive WSI curation of apparent artifactual content and on an interpretability mechanism via a weakly supervised attention based multiple instance learning algorithm that further utilizes clustering to constrain the search space. The automatically identified patterns of high diagnostic value are used to classify the WSI as representative of a short or a long survivor. Identifying tumor morphologic patterns associated with short and long OS will allow the clinical neuropathologist to provide additional prognostic information gleaned during microscopic assessment to the treating team, as well as suggest avenues of biological investigation for understanding and potentially treating glioblastoma.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge