M. Hamed Mozaffari
Spectral unmixing of Raman microscopic images of single human cells using Independent Component Analysis
Oct 25, 2021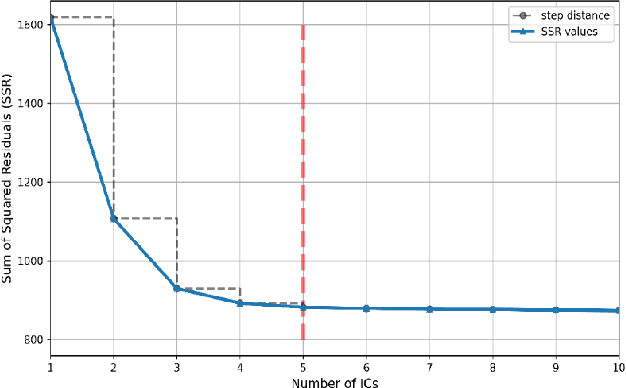
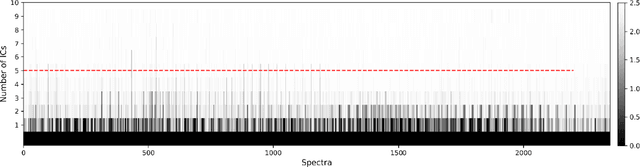
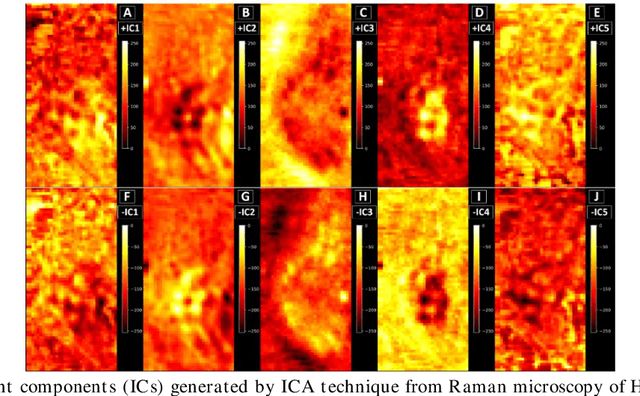
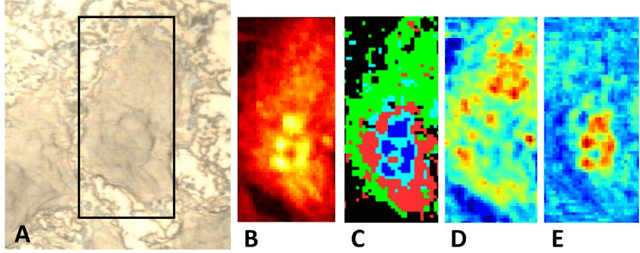
Abstract:Application of independent component analysis (ICA) as an unmixing and image clustering technique for high spatial resolution Raman maps is reported. A hyperspectral map of a fixed human cell was collected by a Raman micro spectrometer in a raster pattern on a 0.5um grid. Unlike previously used unsupervised machine learning techniques such as principal component analysis, ICA is based on non-Gaussianity and statistical independence of data which is the case for mixture Raman spectra. Hence, ICA is a great candidate for assembling pseudo-colour maps from the spectral hypercube of Raman spectra. Our experimental results revealed that ICA is capable of reconstructing false colour maps of Raman hyperspectral data of human cells, showing the nuclear region constituents as well as subcellular organelle in the cytoplasm and distribution of mitochondria in the perinuclear region. Minimum preprocessing requirements and label-free nature of the ICA method make it a great unmixed method for extraction of endmembers in Raman hyperspectral maps of living cells.
Raman spectral analysis of mixtures with one-dimensional convolutional neural network
Jun 01, 2021

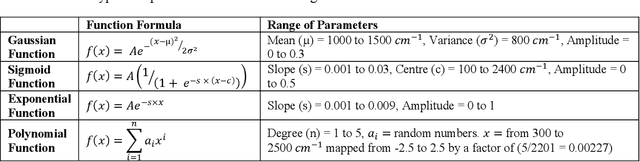
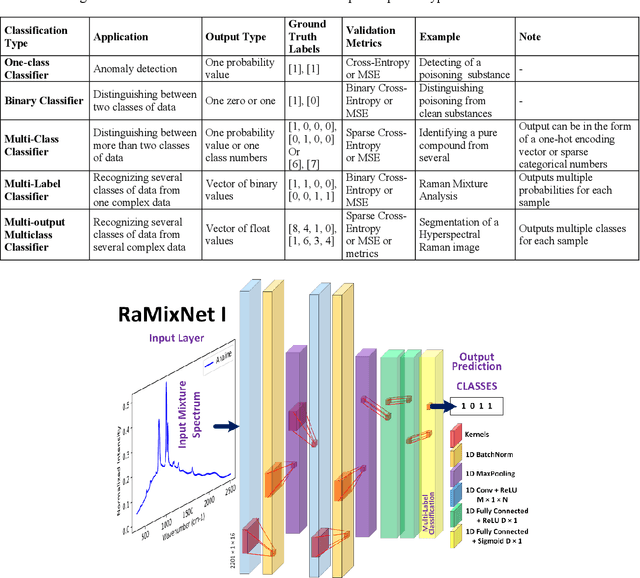
Abstract:Recently, the combination of robust one-dimensional convolutional neural networks (1-D CNNs) and Raman spectroscopy has shown great promise in rapid identification of unknown substances with good accuracy. Using this technique, researchers can recognize a pure compound and distinguish it from unknown substances in a mixture. The novelty of this approach is that the trained neural network operates automatically without any pre- or post-processing of data. Some studies have attempted to extend this technique to the classification of pure compounds in an unknown mixture. However, the application of 1-D CNNs has typically been restricted to binary classifications of pure compounds. Here we will highlight a new approach in spectral recognition and quantification of chemical components in a multicomponent mixture. Two 1-D CNN models, RaMixNet I and II, have been developed for this purpose. The former is for rapid classification of components in a mixture while the latter is for quantitative determination of those constituents. In the proposed method, there is no limit to the number of compounds in a mixture. A data augmentation method is also introduced by adding random baselines to the Raman spectra. The experimental results revealed that the classification accuracy of RaMixNet I and II is 100% for analysis of unknown test mixtures; at the same time, the RaMixNet II model may achieve a regression accuracy of 88% for the quantification of each component.
One-dimensional Active Contour Models for Raman Spectrum Baseline Correction
Apr 26, 2021



Abstract:Raman spectroscopy is a powerful and non-invasive method for analysis of chemicals and detection of unknown substances. However, Raman signal is so weak that background noise can distort the actual Raman signal. These baseline shifts that exist in the Raman spectrum might deteriorate analytical results. In this paper, a modified version of active contour models in one-dimensional space has been proposed for the baseline correction of Raman spectra. Our technique, inspired by principles of physics and heuristic optimization methods, iteratively deforms an initialized curve toward the desired baseline. The performance of the proposed algorithm was evaluated and compared with similar techniques using simulated Raman spectra. The results showed that the 1D active contour model outperforms many iterative baseline correction methods. The proposed algorithm was successfully applied to experimental Raman spectral data, and the results indicate that the baseline of Raman spectra can be automatically subtracted.
PS8-Net: A Deep Convolutional Neural Network to Predict the Eight-State Protein Secondary Structure
Sep 22, 2020



Abstract:Protein secondary structure is crucial to creating an information bridge between the primary and tertiary (3D) structures. Precise prediction of eight-state protein secondary structure (PSS) has significantly utilized in the structural and functional analysis of proteins in bioinformatics. Deep learning techniques have been recently applied in this research area and raised the eight-state (Q8) protein secondary structure prediction accuracy remarkably. Nevertheless, from a theoretical standpoint, there are still lots of rooms for improvement, specifically in the eight-state PSS prediction. In this study, we have presented a new deep convolutional neural network (DCNN), namely PS8-Net, to enhance the accuracy of eight-class PSS prediction. The input of this architecture is a carefully constructed feature matrix from the proteins sequence features and profile features. We introduce a new PS8 module in the network, which is applied with skip connection to extracting the long-term inter-dependencies from higher layers, obtaining local contexts in earlier layers, and achieving global information during secondary structure prediction. Our proposed PS8-Net achieves 76.89%, 71.94%, 76.86%, and 75.26% Q8 accuracy respectively on benchmark CullPdb6133, CB513, CASP10, and CASP11 datasets. This architecture enables the efficient processing of local and global interdependencies between amino acids to make an accurate prediction of each class. To the best of our knowledge, PS8-Net experiment results demonstrate that it outperforms all the state-of-the-art methods on the aforementioned benchmark datasets.
A Review of 1D Convolutional Neural Networks toward Unknown Substance Identification in Portable Raman Spectrometer
Jun 18, 2020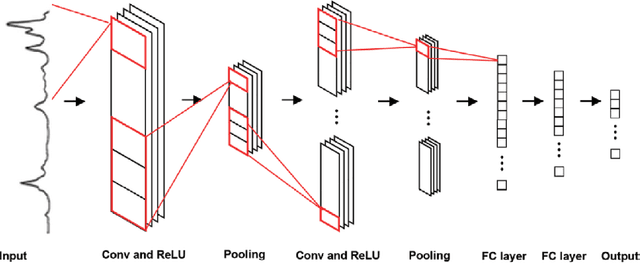



Abstract:Raman spectroscopy is a powerful analytical tool with applications ranging from quality control to cutting edge biomedical research. One particular area which has seen tremendous advances in the past decade is the development of powerful handheld Raman spectrometers. They have been adopted widely by first responders and law enforcement agencies for the field analysis of unknown substances. Field detection and identification of unknown substances with Raman spectroscopy rely heavily on the spectral matching capability of the devices on hand. Conventional spectral matching algorithms (such as correlation, dot product, etc.) have been used in identifying unknown Raman spectrum by comparing the unknown to a large reference database. This is typically achieved through brute-force summation of pixel-by-pixel differences between the reference and the unknown spectrum. Conventional algorithms have noticeable drawbacks. For example, they tend to work well with identifying pure compounds but less so for mixture compounds. For instance, limited reference spectra inaccessible databases with a large number of classes relative to the number of samples have been a setback for the widespread usage of Raman spectroscopy for field analysis applications. State-of-the-art deep learning methods (specifically convolutional neural networks CNNs), as an alternative approach, presents a number of advantages over conventional spectral comparison algorism. With optimization, they are ideal to be deployed in handheld spectrometers for field detection of unknown substances. In this study, we present a comprehensive survey in the use of one-dimensional CNNs for Raman spectrum identification. Specifically, we highlight the use of this powerful deep learning technique for handheld Raman spectrometers taking into consideration the potential limit in power consumption and computation ability of handheld systems.
Deep Learning for Automatic Tracking of Tongue Surface in Real-time Ultrasound Videos, Landmarks instead of Contours
Mar 16, 2020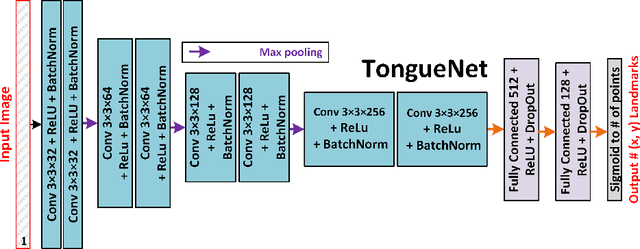
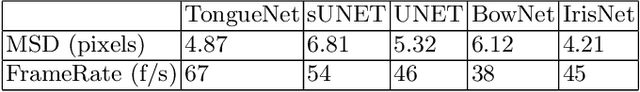

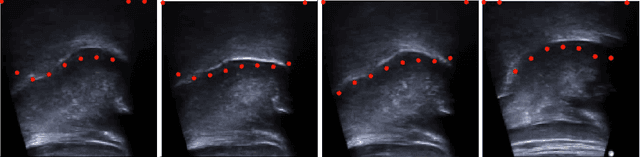
Abstract:One usage of medical ultrasound imaging is to visualize and characterize human tongue shape and motion during a real-time speech to study healthy or impaired speech production. Due to the low-contrast characteristic and noisy nature of ultrasound images, it might require expertise for non-expert users to recognize tongue gestures in applications such as visual training of a second language. Moreover, quantitative analysis of tongue motion needs the tongue dorsum contour to be extracted, tracked, and visualized. Manual tongue contour extraction is a cumbersome, subjective, and error-prone task. Furthermore, it is not a feasible solution for real-time applications. The growth of deep learning has been vigorously exploited in various computer vision tasks, including ultrasound tongue contour tracking. In the current methods, the process of tongue contour extraction comprises two steps of image segmentation and post-processing. This paper presents a new novel approach of automatic and real-time tongue contour tracking using deep neural networks. In the proposed method, instead of the two-step procedure, landmarks of the tongue surface are tracked. This novel idea enables researchers in this filed to benefits from available previously annotated databases to achieve high accuracy results. Our experiment disclosed the outstanding performances of the proposed technique in terms of generalization, performance, and accuracy.
Real-time Ultrasound-enhanced Multimodal Imaging of Tongue using 3D Printable Stabilizer System: A Deep Learning Approach
Nov 22, 2019



Abstract:Despite renewed awareness of the importance of articulation, it remains a challenge for instructors to handle the pronunciation needs of language learners. There are relatively scarce pedagogical tools for pronunciation teaching and learning. Unlike inefficient, traditional pronunciation instructions like listening and repeating, electronic visual feedback (EVF) systems such as ultrasound technology have been employed in new approaches. Recently, an ultrasound-enhanced multimodal method has been developed for visualizing tongue movements of a language learner overlaid on the face-side of the speaker's head. That system was evaluated for several language courses via a blended learning paradigm at the university level. The result was asserted that visualizing the articulator's system as biofeedback to language learners will significantly improve articulation learning efficiency. In spite of the successful usage of multimodal techniques for pronunciation training, it still requires manual works and human manipulation. In this article, we aim to contribute to this growing body of research by addressing difficulties of the previous approaches by proposing a new comprehensive, automatic, real-time multimodal pronunciation training system, benefits from powerful artificial intelligence techniques. The main objective of this research was to combine the advantages of ultrasound technology, three-dimensional printing, and deep learning algorithms to enhance the performance of previous systems. Our preliminary pedagogical evaluation of the proposed system revealed a significant improvement in flexibility, control, robustness, and autonomy.
IrisNet: Deep Learning for Automatic and Real-time Tongue Contour Tracking in Ultrasound Video Data using Peripheral Vision
Nov 10, 2019
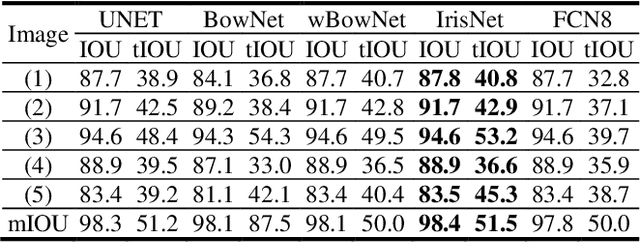


Abstract:The progress of deep convolutional neural networks has been successfully exploited in various real-time computer vision tasks such as image classification and segmentation. Owing to the development of computational units, availability of digital datasets, and improved performance of deep learning models, fully automatic and accurate tracking of tongue contours in real-time ultrasound data became practical only in recent years. Recent studies have shown that the performance of deep learning techniques is significant in the tracking of ultrasound tongue contours in real-time applications such as pronunciation training using multimodal ultrasound-enhanced approaches. Due to the high correlation between ultrasound tongue datasets, it is feasible to have a general model that accomplishes automatic tongue tracking for almost all datasets. In this paper, we proposed a deep learning model comprises of a convolutional module mimicking the peripheral vision ability of the human eye to handle real-time, accurate, and fully automatic tongue contour tracking tasks, applicable for almost all primary ultrasound tongue datasets. Qualitative and quantitative assessment of IrisNet on different ultrasound tongue datasets and PASCAL VOC2012 revealed its outstanding generalization achievement in compare with similar techniques.
Transfer Learning for Ultrasound Tongue Contour Extraction with Different Domains
Jun 10, 2019
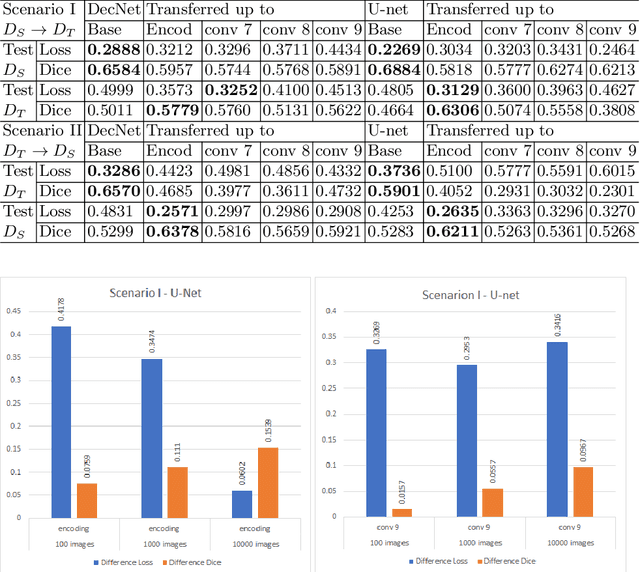

Abstract:Medical ultrasound technology is widely used in routine clinical applications such as disease diagnosis and treatment as well as other applications like real-time monitoring of human tongue shapes and motions as visual feedback in second language training. Due to the low-contrast characteristic and noisy nature of ultrasound images, it might require expertise for non-expert users to recognize tongue gestures. Manual tongue segmentation is a cumbersome, subjective, and error-prone task. Furthermore, it is not a feasible solution for real-time applications. In the last few years, deep learning methods have been used for delineating and tracking tongue dorsum. Deep convolutional neural networks (DCNNs), which have shown to be successful in medical image analysis tasks, are typically weak for the same task on different domains. In many cases, DCNNs trained on data acquired with one ultrasound device, do not perform well on data of varying ultrasound device or acquisition protocol. Domain adaptation is an alternative solution for this difficulty by transferring the weights from the model trained on a large annotated legacy dataset to a new model for adapting on another different dataset using fine-tuning. In this study, after conducting extensive experiments, we addressed the problem of domain adaptation on small ultrasound datasets for tongue contour extraction. We trained a U-net network comprises of an encoder-decoder path from scratch, and then with several surrogate scenarios, some parts of the trained network were fine-tuned on another dataset as the domain-adapted networks. We repeat scenarios from target to source domains to find a balance point for knowledge transfer from source to target and vice versa. The performance of new fine-tuned networks was evaluated on the same task with images from different domains.
BowNet: Dilated Convolution Neural Network for Ultrasound Tongue Contour Extraction
Jun 10, 2019


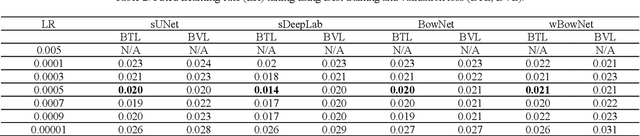
Abstract:Ultrasound imaging is safe, relatively affordable, and capable of real-time performance. One application of this technology is to visualize and to characterize human tongue shape and motion during a real-time speech to study healthy or impaired speech production. Due to the noisy nature of ultrasound images with low-contrast characteristic, it might require expertise for non-expert users to recognize organ shape such as tongue surface (dorsum). To alleviate this difficulty for quantitative analysis of tongue shape and motion, tongue surface can be extracted, tracked, and visualized instead of the whole tongue region. Delineating the tongue surface from each frame is a cumbersome, subjective, and error-prone task. Furthermore, the rapidity and complexity of tongue gestures have made it a challenging task, and manual segmentation is not a feasible solution for real-time applications. Employing the power of state-of-the-art deep neural network models and training techniques, it is feasible to implement new fully-automatic, accurate, and robust segmentation methods with the capability of real-time performance, applicable for tracking of the tongue contours during the speech. This paper presents two novel deep neural network models named BowNet and wBowNet benefits from the ability of global prediction of decoding-encoding models, with integrated multi-scale contextual information, and capability of full-resolution (local) extraction of dilated convolutions. Experimental results using several ultrasound tongue image datasets revealed that the combination of both localization and globalization searching could improve prediction result significantly. Assessment of BowNet models using both qualitatively and quantitatively studies showed them outstanding achievements in terms of accuracy and robustness in comparison with similar techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge