Li-Lin Tay
Spectral unmixing of Raman microscopic images of single human cells using Independent Component Analysis
Oct 25, 2021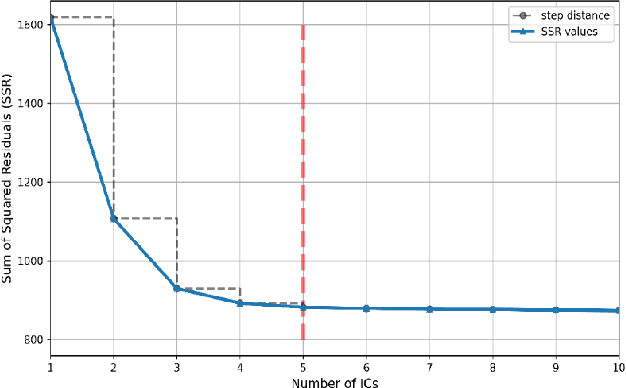
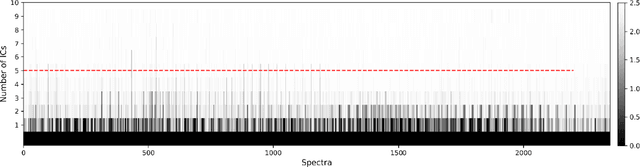
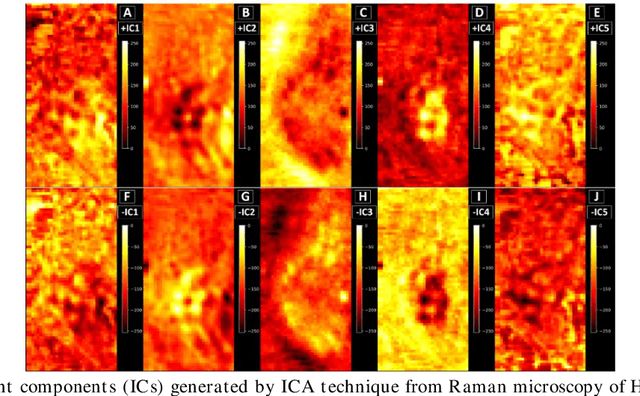
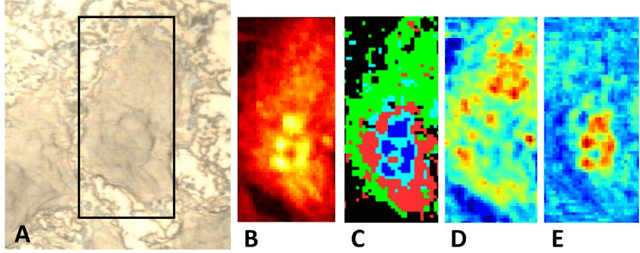
Abstract:Application of independent component analysis (ICA) as an unmixing and image clustering technique for high spatial resolution Raman maps is reported. A hyperspectral map of a fixed human cell was collected by a Raman micro spectrometer in a raster pattern on a 0.5um grid. Unlike previously used unsupervised machine learning techniques such as principal component analysis, ICA is based on non-Gaussianity and statistical independence of data which is the case for mixture Raman spectra. Hence, ICA is a great candidate for assembling pseudo-colour maps from the spectral hypercube of Raman spectra. Our experimental results revealed that ICA is capable of reconstructing false colour maps of Raman hyperspectral data of human cells, showing the nuclear region constituents as well as subcellular organelle in the cytoplasm and distribution of mitochondria in the perinuclear region. Minimum preprocessing requirements and label-free nature of the ICA method make it a great unmixed method for extraction of endmembers in Raman hyperspectral maps of living cells.
Raman spectral analysis of mixtures with one-dimensional convolutional neural network
Jun 01, 2021

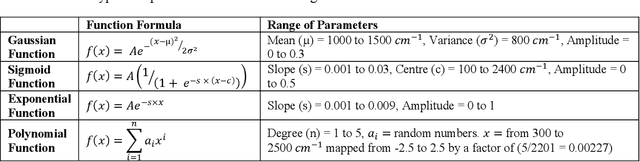
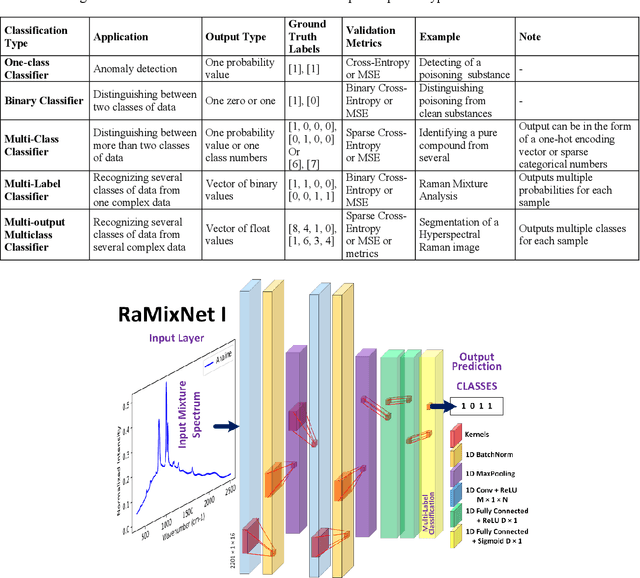
Abstract:Recently, the combination of robust one-dimensional convolutional neural networks (1-D CNNs) and Raman spectroscopy has shown great promise in rapid identification of unknown substances with good accuracy. Using this technique, researchers can recognize a pure compound and distinguish it from unknown substances in a mixture. The novelty of this approach is that the trained neural network operates automatically without any pre- or post-processing of data. Some studies have attempted to extend this technique to the classification of pure compounds in an unknown mixture. However, the application of 1-D CNNs has typically been restricted to binary classifications of pure compounds. Here we will highlight a new approach in spectral recognition and quantification of chemical components in a multicomponent mixture. Two 1-D CNN models, RaMixNet I and II, have been developed for this purpose. The former is for rapid classification of components in a mixture while the latter is for quantitative determination of those constituents. In the proposed method, there is no limit to the number of compounds in a mixture. A data augmentation method is also introduced by adding random baselines to the Raman spectra. The experimental results revealed that the classification accuracy of RaMixNet I and II is 100% for analysis of unknown test mixtures; at the same time, the RaMixNet II model may achieve a regression accuracy of 88% for the quantification of each component.
One-dimensional Active Contour Models for Raman Spectrum Baseline Correction
Apr 26, 2021



Abstract:Raman spectroscopy is a powerful and non-invasive method for analysis of chemicals and detection of unknown substances. However, Raman signal is so weak that background noise can distort the actual Raman signal. These baseline shifts that exist in the Raman spectrum might deteriorate analytical results. In this paper, a modified version of active contour models in one-dimensional space has been proposed for the baseline correction of Raman spectra. Our technique, inspired by principles of physics and heuristic optimization methods, iteratively deforms an initialized curve toward the desired baseline. The performance of the proposed algorithm was evaluated and compared with similar techniques using simulated Raman spectra. The results showed that the 1D active contour model outperforms many iterative baseline correction methods. The proposed algorithm was successfully applied to experimental Raman spectral data, and the results indicate that the baseline of Raman spectra can be automatically subtracted.
A Review of 1D Convolutional Neural Networks toward Unknown Substance Identification in Portable Raman Spectrometer
Jun 18, 2020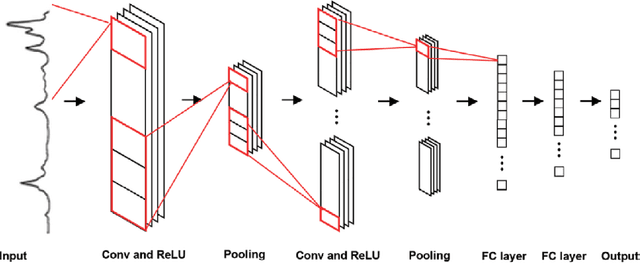



Abstract:Raman spectroscopy is a powerful analytical tool with applications ranging from quality control to cutting edge biomedical research. One particular area which has seen tremendous advances in the past decade is the development of powerful handheld Raman spectrometers. They have been adopted widely by first responders and law enforcement agencies for the field analysis of unknown substances. Field detection and identification of unknown substances with Raman spectroscopy rely heavily on the spectral matching capability of the devices on hand. Conventional spectral matching algorithms (such as correlation, dot product, etc.) have been used in identifying unknown Raman spectrum by comparing the unknown to a large reference database. This is typically achieved through brute-force summation of pixel-by-pixel differences between the reference and the unknown spectrum. Conventional algorithms have noticeable drawbacks. For example, they tend to work well with identifying pure compounds but less so for mixture compounds. For instance, limited reference spectra inaccessible databases with a large number of classes relative to the number of samples have been a setback for the widespread usage of Raman spectroscopy for field analysis applications. State-of-the-art deep learning methods (specifically convolutional neural networks CNNs), as an alternative approach, presents a number of advantages over conventional spectral comparison algorism. With optimization, they are ideal to be deployed in handheld spectrometers for field detection of unknown substances. In this study, we present a comprehensive survey in the use of one-dimensional CNNs for Raman spectrum identification. Specifically, we highlight the use of this powerful deep learning technique for handheld Raman spectrometers taking into consideration the potential limit in power consumption and computation ability of handheld systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge