Lucas C Parra
Full-Head Segmentation of MRI with Abnormal Brain Anatomy: Model and Data Release
Jan 30, 2025



Abstract:The goal of this work was to develop a deep network for whole-head segmentation, including clinical MRIs with abnormal anatomy, and compile the first public benchmark dataset for this purpose. We collected 91 MRIs with volumetric segmentation labels for a diverse set of human subjects (4 normal, 32 traumatic brain injuries, and 57 strokes). These clinical cases are characterized by extended cerebrospinal fluid (CSF) in regions normally containing the brain. Training labels were generated by manually correcting initial automated segmentations for skin/scalp, skull, CSF, gray matter, white matter, air cavity, and extracephalic air. We developed a MultiAxial network consisting of three 2D U-Net models that operate independently in sagittal, axial, and coronal planes and are then combined to produce a single 3D segmentation. The MultiAxial network achieved test-set Dice scores of 0.88 (median plus-minus 0.04). For brain tissue, it significantly outperforms existing brain segmentation methods (MultiAxial: 0.898 plus-minus 0.041, SynthSeg: 0.758 plus-minus 0.054, BrainChop: 0.757 plus-minus 0.125). The MultiAxial network gains in robustness by avoiding the need for coregistration with an atlas. It performed well in regions with abnormal anatomy and on images that have been de-identified. It enables more robust current flow modeling when incorporated into ROAST, a widely-used modeling toolbox for transcranial electric stimulation. We are releasing a state-of-the-art model for whole-head MRI segmentation, along with a dataset of 61 clinical MRIs and training labels, including non-brain structures. Together, the model and data may serve as a benchmark for future efforts.
Recurrent Joint Embedding Predictive Architecture with Recurrent Forward Propagation Learning
Nov 10, 2024Abstract:Conventional computer vision models rely on very deep, feedforward networks processing whole images and trained offline with extensive labeled data. In contrast, biological vision relies on comparatively shallow, recurrent networks that analyze sequences of fixated image patches, learning continuously in real-time without explicit supervision. This work introduces a vision network inspired by these biological principles. Specifically, it leverages a joint embedding predictive architecture incorporating recurrent gated circuits. The network learns by predicting the representation of the next image patch (fixation) based on the sequence of past fixations, a form of self-supervised learning. We show mathematical and empirically that the training algorithm avoids the problem of representational collapse. We also introduce \emph{Recurrent-Forward Propagation}, a learning algorithm that avoids biologically unrealistic backpropagation through time or memory-inefficient real-time recurrent learning. We show mathematically that the algorithm implements exact gradient descent for a large class of recurrent architectures, and confirm empirically that it learns efficiently. This paper focuses on these theoretical innovations and leaves empirical evaluation of performance in downstream tasks, and analysis of representational similarity with biological vision for future work.
Deep learning achieves radiologist-level performance of tumor segmentation in breast MRI
Sep 21, 2020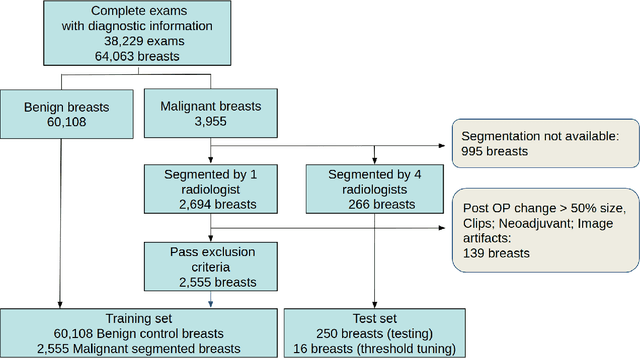
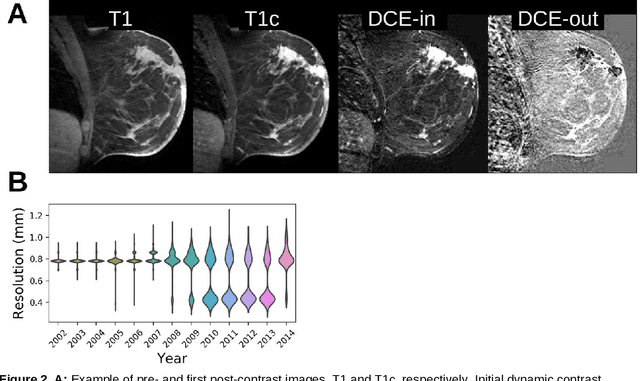
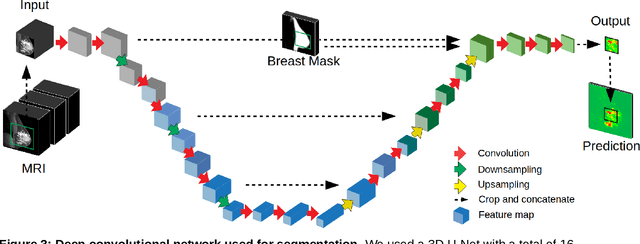
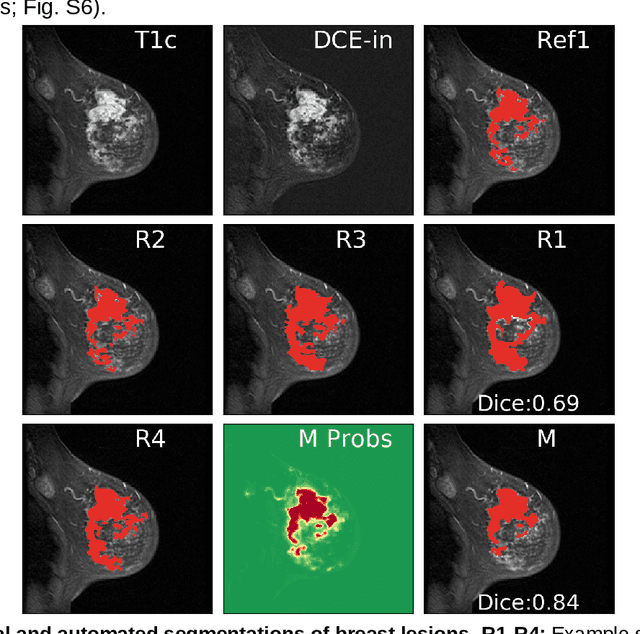
Abstract:Purpose: The goal of this research was to develop a deep network architecture that achieves fully-automated radiologist-level segmentation of breast tumors in MRI. Materials and Methods: We leveraged 38,229 clinical MRI breast exams collected retrospectively from women aged 12-94 (mean age 54) who presented between 2002 and 2014 at a single clinical site. The training set for the network consisted of 2,555 malignant breasts that were segmented in 2D by experienced radiologists, as well as 60,108 benign breasts that served as negative controls. The test set consisted of 250 exams with tumors segmented independently by four radiologists. We selected among several 3D deep convolutional neural network architectures, input modalities and harmonization methods. The outcome measure was the Dice score for 2D segmentation, and was compared between the network and radiologists using the Wilcoxon signed-rank test and the TOST procedure. Results: The best-performing network on the training set was a volumetric U-Net with contrast enhancement dynamic as input and with intensity normalized for each exam. In the test set the median Dice score of this network was 0.77. The performance of the network was equivalent to that of the radiologists (TOST procedure with radiologist performance of 0.69-0.84 as equivalence bounds: p = 5e-10 and p = 2e-5, respectively; N = 250) and compares favorably with published state of the art (0.6-0.77). Conclusion: When trained on a dataset of over 60 thousand breasts, a volumetric U-Net performs as well as expert radiologists at segmenting malignant breast lesions in MRI.
Tissue segmentation with deep 3D networks and spatial priors
May 24, 2019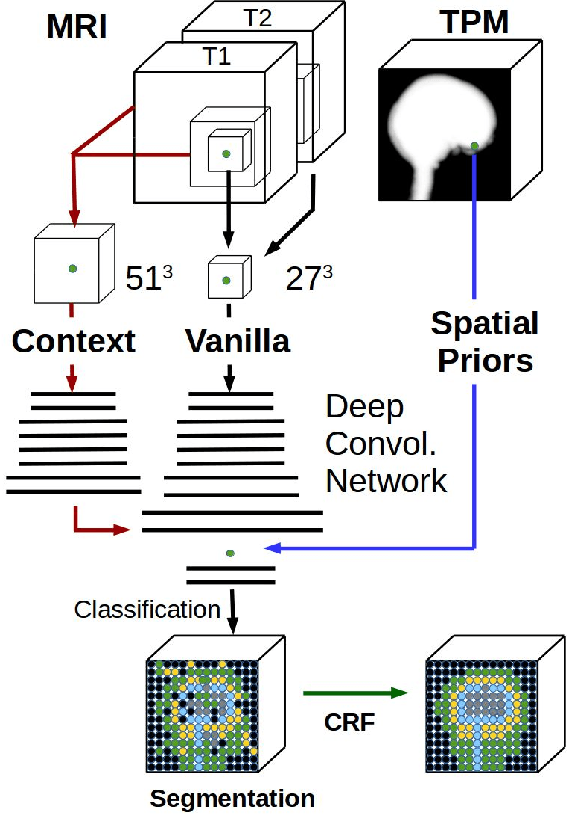
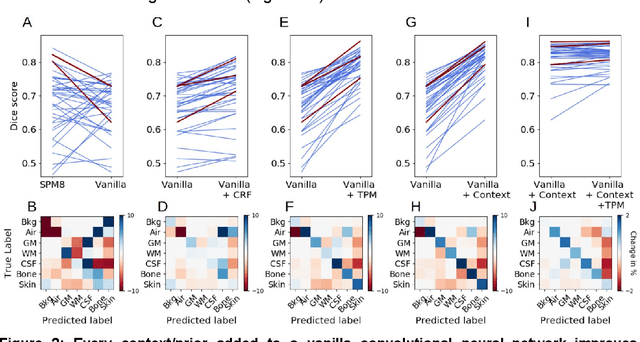

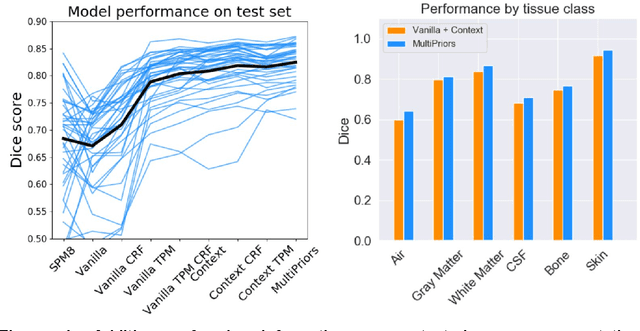
Abstract:Conventional automated segmentation of the human head distinguishes different tissues based on image intensities in an MRI volume and prior tissue probability maps (TPM). This works well for normal head anatomies, but fails in the presence of unexpected lesions. Deep convolutional neural networks leverage instead volumetric spatial patterns and can be trained to segment lesions, but have thus far not integrated prior probabilities. Here we add to a three-dimensional convolutional network spatial priors with a TPM, morphological priors with conditional random fields, and context with a wider field-of-view at lower resolution. The new architecture, which we call MultiPrior, was designed to be a fully-trainable, three-dimensional convolutional network. Thus, the resulting architecture represents a neural network with learnable spatial memories. When trained on a set of stroke patients and healthy subjects, MultiPrior outperforms the state-of-the-art segmentation tools such as DeepMedic and SPM segmentation. The approach is further demonstrated on patients with disorders of consciousness, where we find that cognitive state correlates positively with gray-matter volumes and negatively with the extent of ventricles. We make the code and trained networks freely available to support future clinical research projects.
Multi-set Canonical Correlation Analysis simply explained
Feb 11, 2018Abstract:There are a multitude of methods to perform multi-set correlated component analysis (MCCA), including some that require iterative solutions. The methods differ on the criterion they optimize and the constraints placed on the solutions. This note focuses perhaps on the simplest version, which can be solved in a single step as the eigenvectors of matrix ${\bf D}^{-1} {\bf R}$. Here ${\bf R}$ is the covariance matrix of the concatenated data, and ${\bf D}$ is its block-diagonal. This note shows that this solution maximizes inter-set correlation (ISC) without further constraints. It also relates the solution to a two step procedure, which first whitens each dataset using PCA, and then performs an additional PCA on the concatenated and whitened data. Both these solutions are known, although a clear derivation and simple implementation are hard to find. This short note aims to remedy this.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge