PhD
College of Engineering, Drexel University, Philadelphia, Pennsylvania, USA
Machine learning applications to DNA subsequence and restriction site analysis
Nov 07, 2020



Abstract:Based on the BioBricks standard, restriction synthesis is a novel catabolic iterative DNA synthesis method that utilizes endonucleases to synthesize a query sequence from a reference sequence. In this work, the reference sequence is built from shorter subsequences by classifying them as applicable or inapplicable for the synthesis method using three different machine learning methods: Support Vector Machines (SVMs), random forest, and Convolution Neural Networks (CNNs). Before applying these methods to the data, a series of feature selection, curation, and reduction steps are applied to create an accurate and representative feature space. Following these preprocessing steps, three different pipelines are proposed to classify subsequences based on their nucleotide sequence and other relevant features corresponding to the restriction sites of over 200 endonucleases. The sensitivity using SVMs, random forest, and CNNs are 94.9%, 92.7%, 91.4%, respectively. Moreover, each method scores lower in specificity with SVMs, random forest, and CNNs resulting in 77.4%, 85.7%, and 82.4%, respectively. In addition to analyzing these results, the misclassifications in SVMs and CNNs are investigated. Across these two models, different features with a derived nucleotide specificity visually contribute more to classification compared to other features. This observation is an important factor when considering new nucleotide sensitivity features for future studies.
Deep learning achieves radiologist-level performance of tumor segmentation in breast MRI
Sep 21, 2020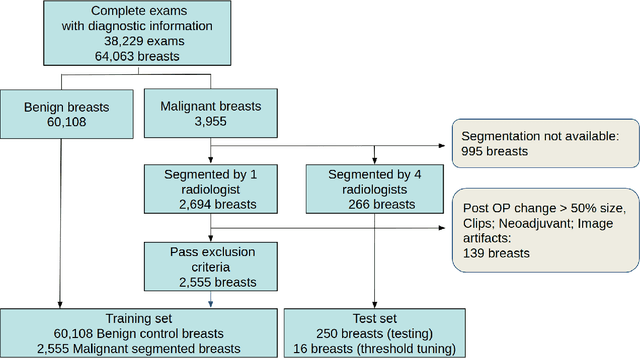
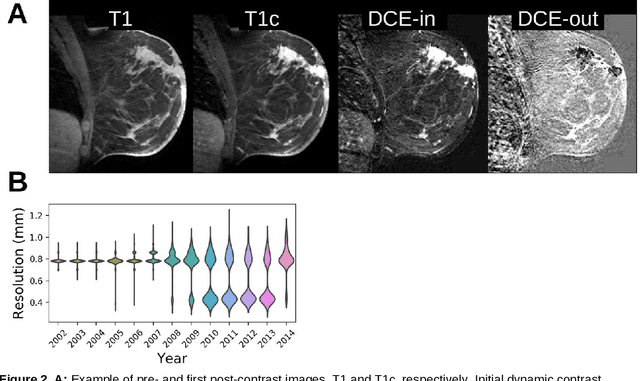
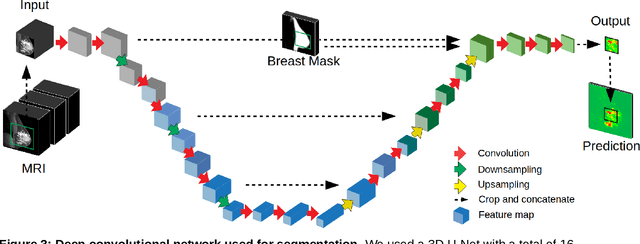
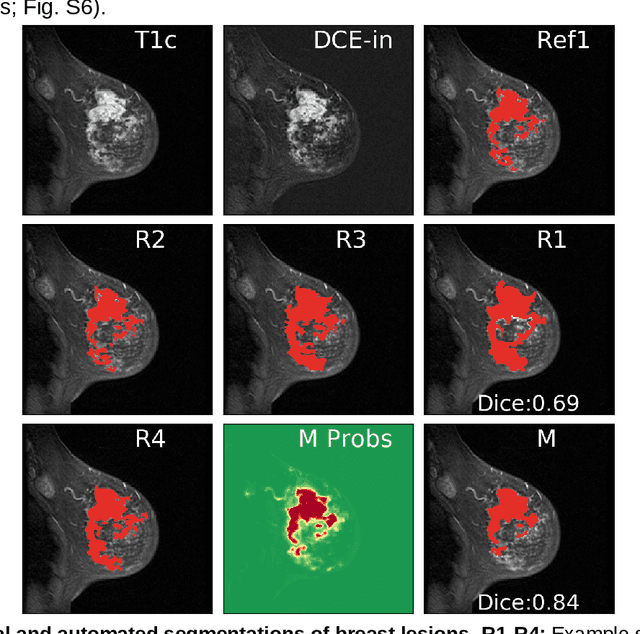
Abstract:Purpose: The goal of this research was to develop a deep network architecture that achieves fully-automated radiologist-level segmentation of breast tumors in MRI. Materials and Methods: We leveraged 38,229 clinical MRI breast exams collected retrospectively from women aged 12-94 (mean age 54) who presented between 2002 and 2014 at a single clinical site. The training set for the network consisted of 2,555 malignant breasts that were segmented in 2D by experienced radiologists, as well as 60,108 benign breasts that served as negative controls. The test set consisted of 250 exams with tumors segmented independently by four radiologists. We selected among several 3D deep convolutional neural network architectures, input modalities and harmonization methods. The outcome measure was the Dice score for 2D segmentation, and was compared between the network and radiologists using the Wilcoxon signed-rank test and the TOST procedure. Results: The best-performing network on the training set was a volumetric U-Net with contrast enhancement dynamic as input and with intensity normalized for each exam. In the test set the median Dice score of this network was 0.77. The performance of the network was equivalent to that of the radiologists (TOST procedure with radiologist performance of 0.69-0.84 as equivalence bounds: p = 5e-10 and p = 2e-5, respectively; N = 250) and compares favorably with published state of the art (0.6-0.77). Conclusion: When trained on a dataset of over 60 thousand breasts, a volumetric U-Net performs as well as expert radiologists at segmenting malignant breast lesions in MRI.
3D Contouring for Breast Tumor in Sonography
Jan 27, 2019



Abstract:Malignant and benign breast tumors present differently in their shape and size on sonography. Morphological information provided by tumor contours are important in clinical diagnosis. However, ultrasound images contain noises and tissue texture; clinical diagnosis thus highly depends on the experience of physicians. The manual way to sketch three-dimensional (3D) contours of breast tumor is a time-consuming and complicate task. If automatic contouring could provide a precise breast tumor contour that might assist physicians in making an accurate diagnosis. This study presents an efficient method for automatically contouring breast tumors in 3D sonography. The proposed method utilizes an efficient segmentation procedure, i.e. level-set method (LSM), to automatic detect contours of breast tumors. This study evaluates 20 cases comprising ten benign and ten malignant tumors. The results of computer simulation reveal that the proposed 3D segmentation method provides robust contouring for breast tumor on ultrasound images. This approach consistently obtains contours similar to those obtained by manual contouring of the breast tumor and can save much of the time required to sketch precise contours.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge