Lotta M. Ellingsen
and for the Alzheimer's Disease Neuroimaging Initiative
Efficient Brain Extraction of MRI Scans with Mild to Moderate Neuropathology
Feb 09, 2026Abstract:Skull stripping magnetic resonance images (MRI) of the human brain is an important process in many image processing techniques, such as automatic segmentation of brain structures. Numerous methods have been developed to perform this task, however, they often fail in the presence of neuropathology and can be inconsistent in defining the boundary of the brain mask. Here, we propose a novel approach to skull strip T1-weighted images in a robust and efficient manner, aiming to consistently segment the outer surface of the brain, including the sulcal cerebrospinal fluid (CSF), while excluding the full extent of the subarachnoid space and meninges. We train a modified version of the U-net on silver-standard ground truth data using a novel loss function based on the signed-distance transform (SDT). We validate our model both qualitatively and quantitatively using held-out data from the training dataset, as well as an independent external dataset. The brain masks used for evaluation partially or fully include the subarachnoid space, which may introduce bias into the comparison; nonetheless, our model demonstrates strong performance on the held-out test data, achieving a consistent mean Dice similarity coefficient (DSC) of 0.964$\pm$0.006 and an average symmetric surface distance (ASSD) of 1.4mm$\pm$0.2mm. Performance on the external dataset is comparable, with a DSC of 0.958$\pm$0.006 and an ASSD of 1.7$\pm$0.2mm. Our method achieves performance comparable to or better than existing state-of-the-art methods for brain extraction, particularly in its highly consistent preservation of the brain's outer surface. The method is publicly available on GitHub.
An Interpretable Vision Transformer as a Fingerprint-Based Diagnostic Aid for Kabuki and Wiedemann-Steiner Syndromes
Feb 06, 2026Abstract:Kabuki syndrome (KS) and Wiedemann-Steiner syndrome (WSS) are rare but distinct developmental disorders that share overlapping clinical features, including neurodevelopmental delay, growth restriction, and persistent fetal fingertip pads. While genetic testing remains the diagnostic gold standard, many individuals with KS or WSS remain undiagnosed due to barriers in access to both genetic testing and expertise. Dermatoglyphic anomalies, despite being established hallmarks of several genetic syndromes, remain an underutilized diagnostic signal in the era of molecular testing. This study presents a vision transformer-based deep learning model that leverages fingerprint images to distinguish individuals with KS and WSS from unaffected controls and from one another. We evaluate model performance across three binary classification tasks. Across the three classification tasks, the model achieved AUC scores of 0.80 (control vs. KS), 0.73 (control vs. WSS), and 0.85 (KS vs. WSS), with corresponding F1 scores of 0.71, 0.72, and 0.83, respectively. Beyond classification, we apply attention-based visualizations to identify fingerprint regions most salient to model predictions, enhancing interpretability. Together, these findings suggest the presence of syndrome-specific fingerprint features, demonstrating the feasibility of a fingerprint-based artificial intelligence (AI) tool as a noninvasive, interpretable, and accessible future diagnostic aid for the early diagnosis of underdiagnosed genetic syndromes.
A Hybrid CNN and ML Framework for Multi-modal Classification of Movement Disorders Using MRI and Brain Structural Features
Feb 05, 2026Abstract:Atypical Parkinsonian Disorders (APD), also known as Parkinson-plus syndrome, are a group of neurodegenerative diseases that include progressive supranuclear palsy (PSP) and multiple system atrophy (MSA). In the early stages, overlapping clinical features often lead to misdiagnosis as Parkinson's disease (PD). Identifying reliable imaging biomarkers for early differential diagnosis remains a critical challenge. In this study, we propose a hybrid framework combining convolutional neural networks (CNNs) with machine learning (ML) techniques to classify APD subtypes versus PD and distinguish between the subtypes themselves: PSP vs. PD, MSA vs. PD, and PSP vs. MSA. The model leverages multi-modal input data, including T1-weighted magnetic resonance imaging (MRI), segmentation masks of 12 deep brain structures associated with APD, and their corresponding volumetric measurements. By integrating these complementary modalities, including image data, structural segmentation masks, and quantitative volume features, the hybrid approach achieved promising classification performance with area under the curve (AUC) scores of 0.95 for PSP vs. PD, 0.86 for MSA vs. PD, and 0.92 for PSP vs. MSA. These results highlight the potential of combining spatial and structural information for robust subtype differentiation. In conclusion, this study demonstrates that fusing CNN-based image features with volume-based ML inputs improves classification accuracy for APD subtypes. The proposed approach may contribute to more reliable early-stage diagnosis, facilitating timely and targeted interventions in clinical practice.
Region-based U-net for accelerated training and enhanced precision in deep brain segmentation
Mar 14, 2024


Abstract:Segmentation of brain structures on MRI is the primary step for further quantitative analysis of brain diseases. Manual segmentation is still considered the gold standard in terms of accuracy; however, such data is extremely time-consuming to generate. This paper presents a deep learning-based segmentation approach for 12 deep-brain structures, utilizing multiple region-based U-Nets. The brain is divided into three focal regions of interest that encompass the brainstem, the ventricular system, and the striatum. Next, three region-based U-nets are run in parallel to parcellate these larger structures into their respective four substructures. This approach not only greatly reduces the training and processing times but also significantly enhances the segmentation accuracy, compared to segmenting the entire MRI image at once. Our approach achieves remarkable accuracy with an average Dice Similarity Coefficient (DSC) of 0.901 and 95% Hausdorff Distance (HD95) of 1.155 mm. The method was compared with state-of-the-art segmentation approaches, demonstrating a high level of accuracy and robustness of the proposed method.
Translating the future: Image-to-image translation for the prediction of future brain metabolism
Feb 06, 2024Abstract:Alzheimer's disease (AD) is a progressive neurodegenerative disorder leading to cognitive decline. [$^{18}$F]-Fluorodeoxyglucose positron emission tomography ([$^{18}$F]-FDG PET) is used to monitor brain metabolism, aiding in the diagnosis and assessment of AD over time. However, the feasibility of multi-time point [$^{18}$F]-FDG PET scans for diagnosis is limited due to radiation exposure, cost, and patient burden. To address this, we have developed a predictive image-to-image translation (I2I) model to forecast future [$^{18}$F]-FDG PET scans using baseline and year-one data. The proposed model employs a convolutional neural network architecture with long-short term memory and was trained on [$^{18}$F]-FDG PET data from 161 individuals from the Alzheimer's Disease Neuroimaging Initiative. Our I2I network showed high accuracy in predicting year-two [18F]-FDG PET scans, with a mean absolute error of 0.031 and a structural similarity index of 0.961. Furthermore, the model successfully predicted PET scans up to seven years post-baseline. Notably, the predicted [$^{18}$F]-FDG PET signal in an AD-susceptible meta-region was highly accurate for individuals with mild cognitive impairment across years. In contrast, a linear model was sufficient for predicting brain metabolism in cognitively normal and dementia subjects. In conclusion, both the I2I network and the linear model could offer valuable prognostic insights, guiding early intervention strategies to preemptively address anticipated declines in brain metabolism and potentially to monitor treatment effects.
Automated brainstem parcellation using multi-atlas segmentation and deep neural network
Feb 05, 2021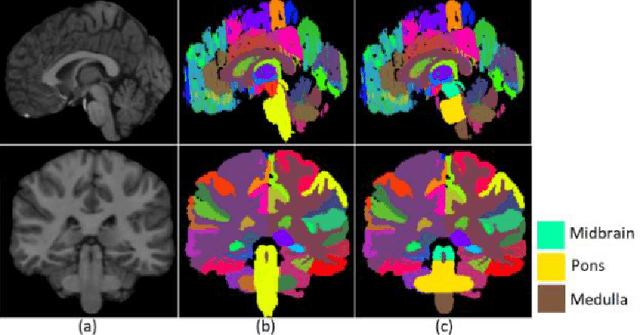
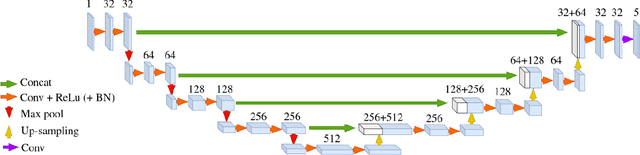
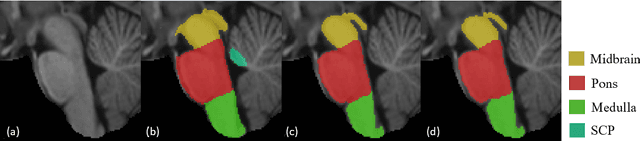
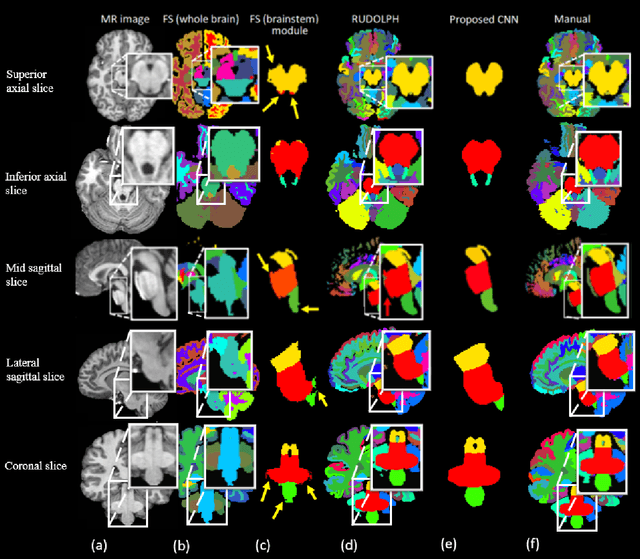
Abstract:About 5-8% of individuals over the age of 60 have dementia. With our ever-aging population this number is likely to increase, making dementia one of the most important threats to public health in the 21st century. Given the phenotypic overlap of individual dementias the diagnosis of dementia is a major clinical challenge, even with current gold standard diagnostic approaches. However, it has been shown that certain dementias show specific structural characteristics in the brain. Progressive supranuclear palsy (PSP) and multiple system atrophy (MSA) are prototypical examples of this phenomenon, as they often present with characteristic brainstem atrophy. More detailed characterization of brain atrophy due to individual diseases is urgently required to select biomarkers and therapeutic targets that are meaningful to each disease. Here we present a joint multi-atlas-segmentation and deep-learning-based segmentation method for fast and robust parcellation of the brainstem into its four sub-structures, i.e., the midbrain, pons, medulla, and superior cerebellar peduncles (SCP), that in turn can provide detailed volumetric information on the brainstem sub-structures affected in PSP and MSA. The method may also benefit other neurodegenerative diseases, such as Parkinson's disease; a condition which is often considered in the differential diagnosis of PSP and MSA. Comparison with state-of-the-art labeling techniques evaluated on ground truth manual segmentations demonstrate that our method is significantly faster than prior methods as well as showing improvement in labeling the brainstem indicating that this strategy may be a viable option to provide a better characterization of the brainstem atrophy seen in PSP and MSA.
Unsupervised brain lesion segmentation from MRI using a convolutional autoencoder
Nov 23, 2018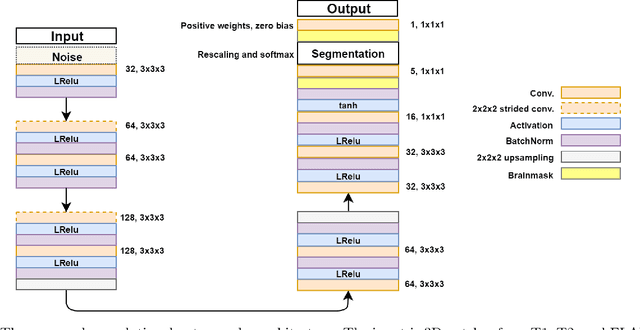
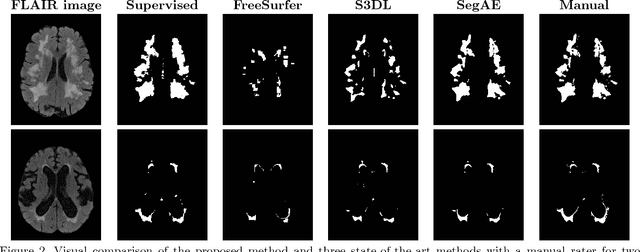

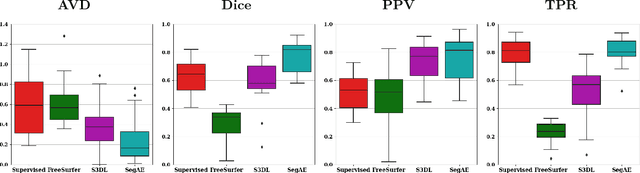
Abstract:Lesions that appear hyperintense in both Fluid Attenuated Inversion Recovery (FLAIR) and T2-weighted magnetic resonance images (MRIs) of the human brain are common in the brains of the elderly population and may be caused by ischemia or demyelination. Lesions are biomarkers for various neurodegenerative diseases, making accurate quantification of them important for both disease diagnosis and progression. Automatic lesion detection using supervised learning requires manually annotated images, which can often be impractical to acquire. Unsupervised lesion detection, on the other hand, does not require any manual delineation; however, these methods can be challenging to construct due to the variability in lesion load, placement of lesions, and voxel intensities. Here we present a novel approach to address this problem using a convolutional autoencoder, which learns to segment brain lesions as well as the white matter, gray matter, and cerebrospinal fluid by reconstructing FLAIR images as conical combinations of softmax layer outputs generated from the corresponding T1, T2, and FLAIR images. Some of the advantages of this model are that it accurately learns to segment lesions regardless of lesion load, and it can be used to quickly and robustly segment new images that were not in the training set. Comparisons with state-of-the-art segmentation methods evaluated on ground truth manual labels indicate that the proposed method works well for generating accurate lesion segmentations without the need for manual annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge