Magnus Magnusson
and for the Alzheimer's Disease Neuroimaging Initiative
Region-based U-net for accelerated training and enhanced precision in deep brain segmentation
Mar 14, 2024


Abstract:Segmentation of brain structures on MRI is the primary step for further quantitative analysis of brain diseases. Manual segmentation is still considered the gold standard in terms of accuracy; however, such data is extremely time-consuming to generate. This paper presents a deep learning-based segmentation approach for 12 deep-brain structures, utilizing multiple region-based U-Nets. The brain is divided into three focal regions of interest that encompass the brainstem, the ventricular system, and the striatum. Next, three region-based U-nets are run in parallel to parcellate these larger structures into their respective four substructures. This approach not only greatly reduces the training and processing times but also significantly enhances the segmentation accuracy, compared to segmenting the entire MRI image at once. Our approach achieves remarkable accuracy with an average Dice Similarity Coefficient (DSC) of 0.901 and 95% Hausdorff Distance (HD95) of 1.155 mm. The method was compared with state-of-the-art segmentation approaches, demonstrating a high level of accuracy and robustness of the proposed method.
Automated brainstem parcellation using multi-atlas segmentation and deep neural network
Feb 05, 2021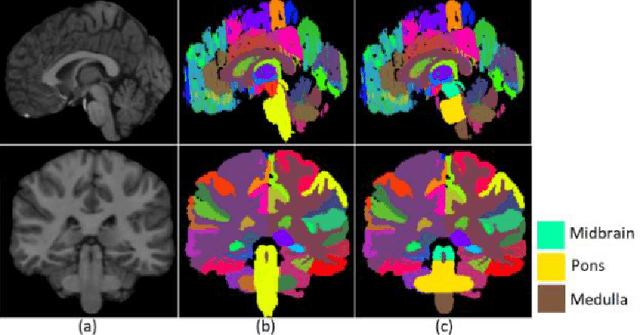
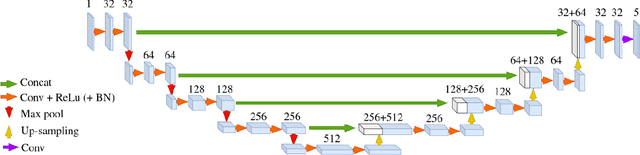
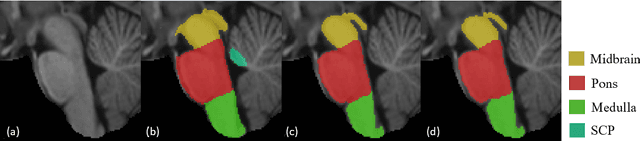
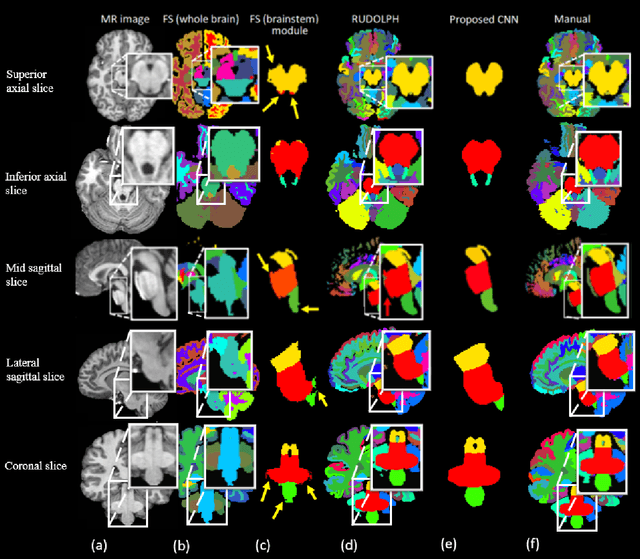
Abstract:About 5-8% of individuals over the age of 60 have dementia. With our ever-aging population this number is likely to increase, making dementia one of the most important threats to public health in the 21st century. Given the phenotypic overlap of individual dementias the diagnosis of dementia is a major clinical challenge, even with current gold standard diagnostic approaches. However, it has been shown that certain dementias show specific structural characteristics in the brain. Progressive supranuclear palsy (PSP) and multiple system atrophy (MSA) are prototypical examples of this phenomenon, as they often present with characteristic brainstem atrophy. More detailed characterization of brain atrophy due to individual diseases is urgently required to select biomarkers and therapeutic targets that are meaningful to each disease. Here we present a joint multi-atlas-segmentation and deep-learning-based segmentation method for fast and robust parcellation of the brainstem into its four sub-structures, i.e., the midbrain, pons, medulla, and superior cerebellar peduncles (SCP), that in turn can provide detailed volumetric information on the brainstem sub-structures affected in PSP and MSA. The method may also benefit other neurodegenerative diseases, such as Parkinson's disease; a condition which is often considered in the differential diagnosis of PSP and MSA. Comparison with state-of-the-art labeling techniques evaluated on ground truth manual segmentations demonstrate that our method is significantly faster than prior methods as well as showing improvement in labeling the brainstem indicating that this strategy may be a viable option to provide a better characterization of the brainstem atrophy seen in PSP and MSA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge