Lorenzo Rosset
adabmDCA 2.0 -- a flexible but easy-to-use package for Direct Coupling Analysis
Jan 30, 2025Abstract:In this methods article, we provide a flexible but easy-to-use implementation of Direct Coupling Analysis (DCA) based on Boltzmann machine learning, together with a tutorial on how to use it. The package \texttt{adabmDCA 2.0} is available in different programming languages (C++, Julia, Python) usable on different architectures (single-core and multi-core CPU, GPU) using a common front-end interface. In addition to several learning protocols for dense and sparse generative DCA models, it allows to directly address common downstream tasks like residue-residue contact prediction, mutational-effect prediction, scoring of sequence libraries and generation of artificial sequences for sequence design. It is readily applicable to protein and RNA sequence data.
Fast, accurate training and sampling of Restricted Boltzmann Machines
May 24, 2024Abstract:Thanks to their simple architecture, Restricted Boltzmann Machines (RBMs) are powerful tools for modeling complex systems and extracting interpretable insights from data. However, training RBMs, as other energy-based models, on highly structured data poses a major challenge, as effective training relies on mixing the Markov chain Monte Carlo simulations used to estimate the gradient. This process is often hindered by multiple second-order phase transitions and the associated critical slowdown. In this paper, we present an innovative method in which the principal directions of the dataset are integrated into a low-rank RBM through a convex optimization procedure. This approach enables efficient sampling of the equilibrium measure via a static Monte Carlo process. By starting the standard training process with a model that already accurately represents the main modes of the data, we bypass the initial phase transitions. Our results show that this strategy successfully trains RBMs to capture the full diversity of data in datasets where previous methods fail. Furthermore, we use the training trajectories to propose a new sampling method, {\em parallel trajectory tempering}, which allows us to sample the equilibrium measure of the trained model much faster than previous optimized MCMC approaches and a better estimation of the log-likelihood. We illustrate the success of the training method on several highly structured datasets.
Fast and Functional Structured Data Generators Rooted in Out-of-Equilibrium Physics
Jul 13, 2023Abstract:In this study, we address the challenge of using energy-based models to produce high-quality, label-specific data in complex structured datasets, such as population genetics, RNA or protein sequences data. Traditional training methods encounter difficulties due to inefficient Markov chain Monte Carlo mixing, which affects the diversity of synthetic data and increases generation times. To address these issues, we use a novel training algorithm that exploits non-equilibrium effects. This approach, applied on the Restricted Boltzmann Machine, improves the model's ability to correctly classify samples and generate high-quality synthetic data in only a few sampling steps. The effectiveness of this method is demonstrated by its successful application to four different types of data: handwritten digits, mutations of human genomes classified by continental origin, functionally characterized sequences of an enzyme protein family, and homologous RNA sequences from specific taxonomies.
Unsupervised hierarchical clustering using the learning dynamics of RBMs
Feb 06, 2023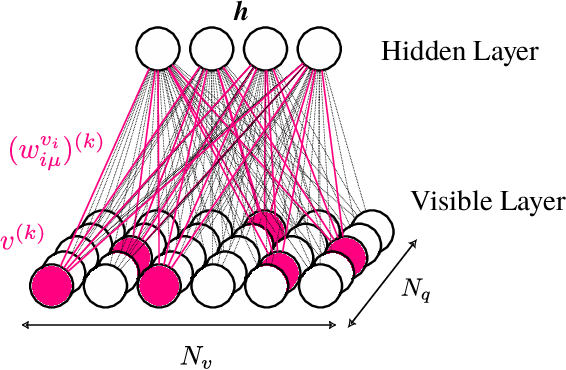
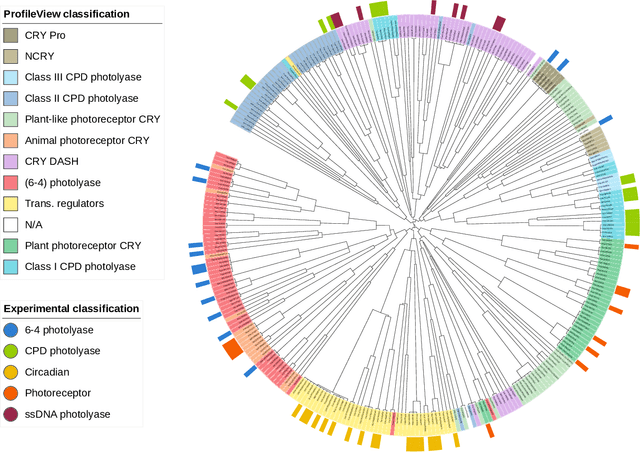
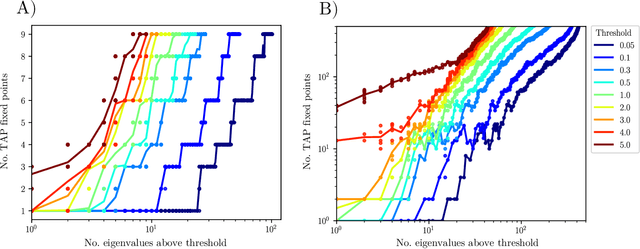
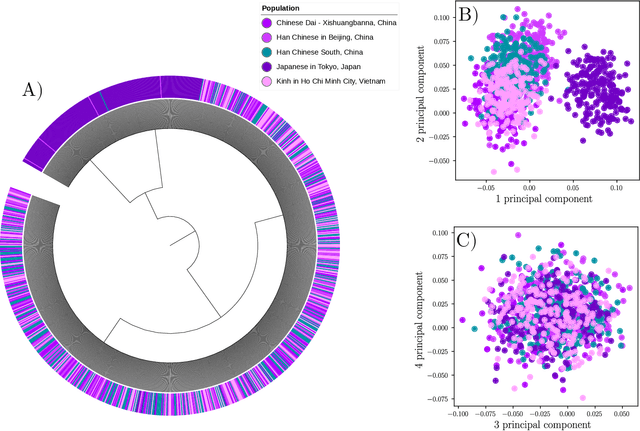
Abstract:Datasets in the real world are often complex and to some degree hierarchical, with groups and sub-groups of data sharing common characteristics at different levels of abstraction. Understanding and uncovering the hidden structure of these datasets is an important task that has many practical applications. To address this challenge, we present a new and general method for building relational data trees by exploiting the learning dynamics of the Restricted Boltzmann Machine (RBM). Our method is based on the mean-field approach, derived from the Plefka expansion, and developed in the context of disordered systems. It is designed to be easily interpretable. We tested our method in an artificially created hierarchical dataset and on three different real-world datasets (images of digits, mutations in the human genome, and a homologous family of proteins). The method is able to automatically identify the hierarchical structure of the data. This could be useful in the study of homologous protein sequences, where the relationships between proteins are critical for understanding their function and evolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge