Lena Podina
Catalyst GFlowNet for electrocatalyst design: A hydrogen evolution reaction case study
Oct 02, 2025Abstract:Efficient and inexpensive energy storage is essential for accelerating the adoption of renewable energy and ensuring a stable supply, despite fluctuations in sources such as wind and solar. Electrocatalysts play a key role in hydrogen energy storage (HES), allowing the energy to be stored as hydrogen. However, the development of affordable and high-performance catalysts for this process remains a significant challenge. We introduce Catalyst GFlowNet, a generative model that leverages machine learning-based predictors of formation and adsorption energy to design crystal surfaces that act as efficient catalysts. We demonstrate the performance of the model through a proof-of-concept application to the hydrogen evolution reaction, a key reaction in HES, for which we successfully identified platinum as the most efficient known catalyst. In future work, we aim to extend this approach to the oxygen evolution reaction, where current optimal catalysts are expensive metal oxides, and open the search space to discover new materials. This generative modeling framework offers a promising pathway for accelerating the search for novel and efficient catalysts.
Enhancing Symbolic Regression and Universal Physics-Informed Neural Networks with Dimensional Analysis
Nov 24, 2024Abstract:We present a new method for enhancing symbolic regression for differential equations via dimensional analysis, specifically Ipsen's and Buckingham pi methods. Since symbolic regression often suffers from high computational costs and overfitting, non-dimensionalizing datasets reduces the number of input variables, simplifies the search space, and ensures that derived equations are physically meaningful. As our main contribution, we integrate Ipsen's method of dimensional analysis with Universal Physics-Informed Neural Networks. We also combine dimensional analysis with the AI Feynman symbolic regression algorithm to show that dimensional analysis significantly improves the accuracy of the recovered equation. The results demonstrate that transforming data into a dimensionless form significantly decreases computation time and improves accuracy of the recovered hidden term. For algebraic equations, using the Buckingham pi theorem reduced complexity, allowing the AI Feynman model to converge faster with fewer data points and lower error rates. For differential equations, Ipsen's method was combined with Universal Physics-Informed Neural Networks (UPINNs) to identify hidden terms more effectively. These findings suggest that integrating dimensional analysis with symbolic regression can significantly lower computational costs, enhance model interpretability, and increase accuracy, providing a robust framework for automated discovery of governing equations in complex systems when data is limited.
Conformalized Physics-Informed Neural Networks
May 13, 2024



Abstract:Physics-informed neural networks (PINNs) are an influential method of solving differential equations and estimating their parameters given data. However, since they make use of neural networks, they provide only a point estimate of differential equation parameters, as well as the solution at any given point, without any measure of uncertainty. Ensemble and Bayesian methods have been previously applied to quantify the uncertainty of PINNs, but these methods may require making strong assumptions on the data-generating process, and can be computationally expensive. Here, we introduce Conformalized PINNs (C-PINNs) that, without making any additional assumptions, utilize the framework of conformal prediction to quantify the uncertainty of PINNs by providing intervals that have finite-sample, distribution-free statistical validity.
Learning Chemotherapy Drug Action via Universal Physics-Informed Neural Networks
Apr 11, 2024Abstract:Quantitative systems pharmacology (QSP) is widely used to assess drug effects and toxicity before the drug goes to clinical trial. However, significant manual distillation of the literature is needed in order to construct a QSP model. Parameters may need to be fit, and simplifying assumptions of the model need to be made. In this work, we apply Universal Physics-Informed Neural Networks (UPINNs) to learn unknown components of various differential equations that model chemotherapy pharmacodynamics. We learn three commonly employed chemotherapeutic drug actions (log-kill, Norton-Simon, and E_max) from synthetic data. Then, we use the UPINN method to fit the parameters for several synthetic datasets simultaneously. Finally, we learn the net proliferation rate in a model of doxorubicin (a chemotherapeutic) pharmacodynamics. As these are only toy examples, we highlight the usefulness of UPINNs in learning unknown terms in pharmacodynamic and pharmacokinetic models.
Denoising Diffusion Restoration Tackles Forward and Inverse Problems for the Laplace Operator
Feb 14, 2024



Abstract:Diffusion models have emerged as a promising class of generative models that map noisy inputs to realistic images. More recently, they have been employed to generate solutions to partial differential equations (PDEs). However, they still struggle with inverse problems in the Laplacian operator, for instance, the Poisson equation, because the eigenvalues that are large in magnitude amplify the measurement noise. This paper presents a novel approach for the inverse and forward solution of PDEs through the use of denoising diffusion restoration models (DDRM). DDRMs were used in linear inverse problems to restore original clean signals by exploiting the singular value decomposition (SVD) of the linear operator. Equivalently, we present an approach to restore the solution and the parameters in the Poisson equation by exploiting the eigenvalues and the eigenfunctions of the Laplacian operator. Our results show that using denoising diffusion restoration significantly improves the estimation of the solution and parameters. Our research, as a result, pioneers the integration of diffusion models with the principles of underlying physics to solve PDEs.
A PINN Approach to Symbolic Differential Operator Discovery with Sparse Data
Dec 09, 2022Abstract:Given ample experimental data from a system governed by differential equations, it is possible to use deep learning techniques to construct the underlying differential operators. In this work we perform symbolic discovery of differential operators in a situation where there is sparse experimental data. This small data regime in machine learning can be made tractable by providing our algorithms with prior information about the underlying dynamics. Physics Informed Neural Networks (PINNs) have been very successful in this regime (reconstructing entire ODE solutions using only a single point or entire PDE solutions with very few measurements of the initial condition). We modify the PINN approach by adding a neural network that learns a representation of unknown hidden terms in the differential equation. The algorithm yields both a surrogate solution to the differential equation and a black-box representation of the hidden terms. These hidden term neural networks can then be converted into symbolic equations using symbolic regression techniques like AI Feynman. In order to achieve convergence of these neural networks, we provide our algorithms with (noisy) measurements of both the initial condition as well as (synthetic) experimental data obtained at later times. We demonstrate strong performance of this approach even when provided with very few measurements of noisy data in both the ODE and PDE regime.
Better Peer Grading through Bayesian Inference
Sep 02, 2022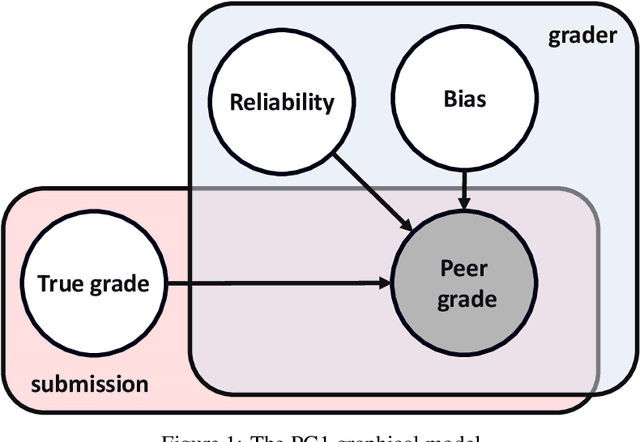
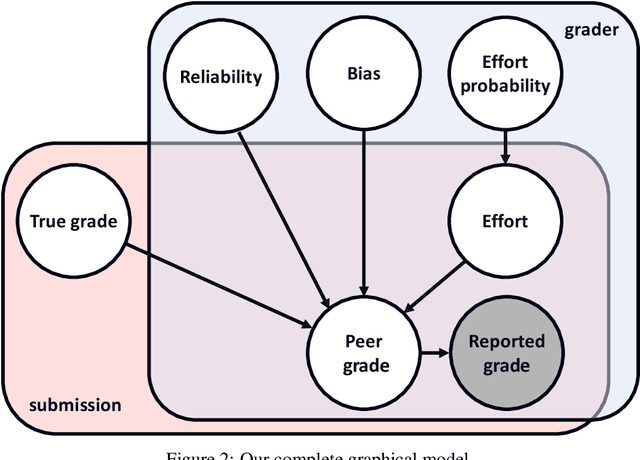
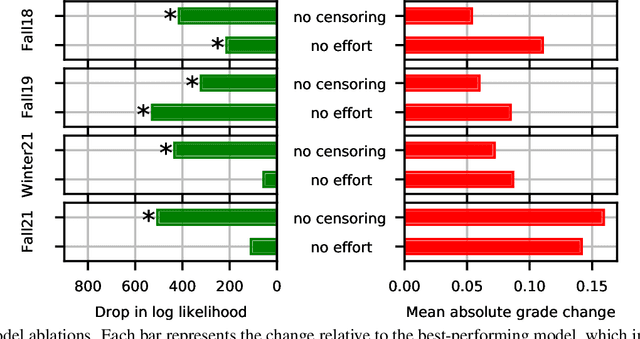
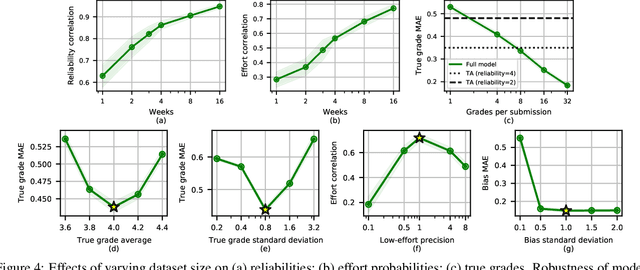
Abstract:Peer grading systems aggregate noisy reports from multiple students to approximate a true grade as closely as possible. Most current systems either take the mean or median of reported grades; others aim to estimate students' grading accuracy under a probabilistic model. This paper extends the state of the art in the latter approach in three key ways: (1) recognizing that students can behave strategically (e.g., reporting grades close to the class average without doing the work); (2) appropriately handling censored data that arises from discrete-valued grading rubrics; and (3) using mixed integer programming to improve the interpretability of the grades assigned to students. We show how to make Bayesian inference practical in this model and evaluate our approach on both synthetic and real-world data obtained by using our implemented system in four large classes. These extensive experiments show that grade aggregation using our model accurately estimates true grades, students' likelihood of submitting uninformative grades, and the variation in their inherent grading error; we also characterize our models' robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge