Kyle Kelley
Bayesian Active Learning for Scanning Probe Microscopy: from Gaussian Processes to Hypothesis Learning
May 30, 2022



Abstract:Recent progress in machine learning methods, and the emerging availability of programmable interfaces for scanning probe microscopes (SPMs), have propelled automated and autonomous microscopies to the forefront of attention of the scientific community. However, enabling automated microscopy requires the development of task-specific machine learning methods, understanding the interplay between physics discovery and machine learning, and fully defined discovery workflows. This, in turn, requires balancing the physical intuition and prior knowledge of the domain scientist with rewards that define experimental goals and machine learning algorithms that can translate these to specific experimental protocols. Here, we discuss the basic principles of Bayesian active learning and illustrate its applications for SPM. We progress from the Gaussian Process as a simple data-driven method and Bayesian inference for physical models as an extension of physics-based functional fits to more complex deep kernel learning methods, structured Gaussian Processes, and hypothesis learning. These frameworks allow for the use of prior data, the discovery of specific functionalities as encoded in spectral data, and exploration of physical laws manifesting during the experiment. The discussed framework can be universally applied to all techniques combining imaging and spectroscopy, SPM methods, nanoindentation, electron microscopy and spectroscopy, and chemical imaging methods, and can be particularly impactful for destructive or irreversible measurements.
Decoding the shift-invariant data: applications for band-excitation scanning probe microscopy
Apr 20, 2021
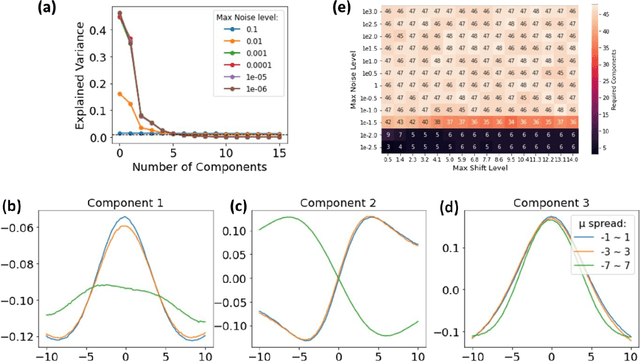


Abstract:A shift-invariant variational autoencoder (shift-VAE) is developed as an unsupervised method for the analysis of spectral data in the presence of shifts along the parameter axis, disentangling the physically-relevant shifts from other latent variables. Using synthetic data sets, we show that the shift-VAE latent variables closely match the ground truth parameters. The shift VAE is extended towards the analysis of band-excitation piezoresponse force microscopy (BE-PFM) data, disentangling the resonance frequency shifts from the peak shape parameters in a model-free unsupervised manner. The extensions of this approach towards denoising of data and model-free dimensionality reduction in imaging and spectroscopic data are further demonstrated. This approach is universal and can also be extended to analysis of X-ray diffraction, photoluminescence, Raman spectra, and other data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge